验证数据展示
经过测试的应用
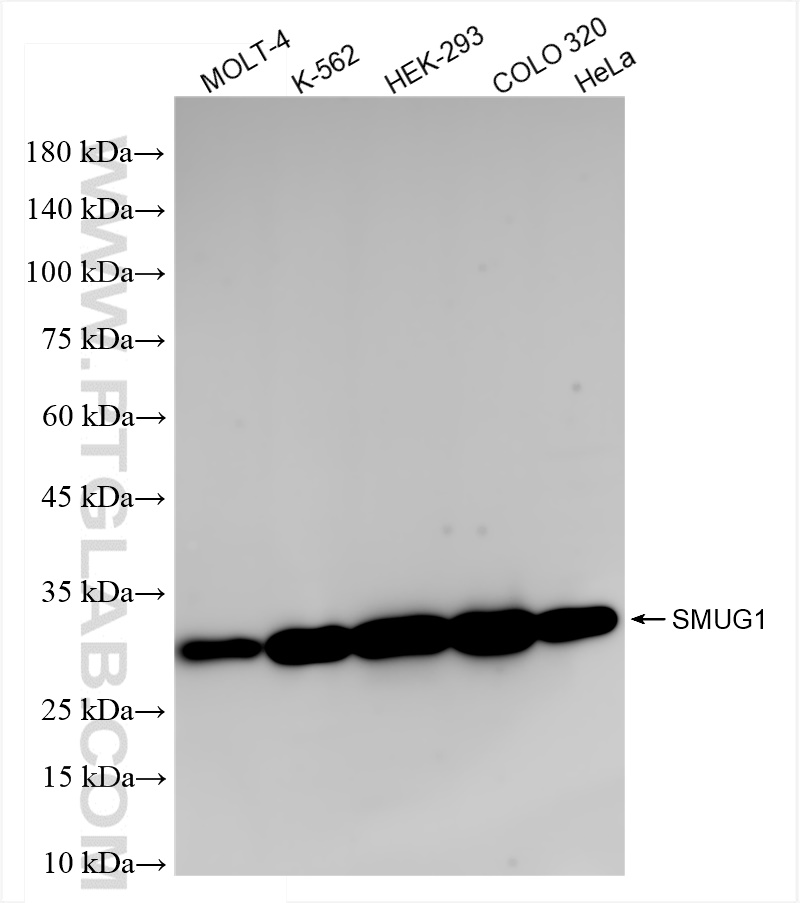
| Positive WB detected in | MOLT-4 cells, K-562 cells, HEK-293 cells, COLO 320 cells, HeLa cells |
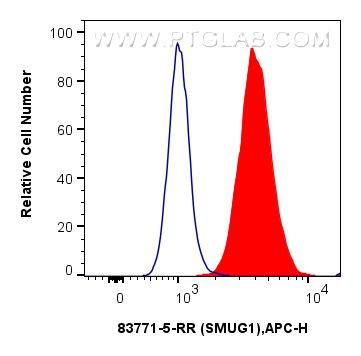
| Positive FC (Intra) detected in | U-2 OS |
推荐稀释比
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.25 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
83771-5-RR targets SMUG1 in WB, FC (Intra), ELISA applications and shows reactivity with human samples.
| Tested Applications | WB, FC (Intra), ELISA Application Description |
| Tested Reactivity | human |
| Immunogen | SMUG1 fusion protein Ag13934 种属同源性预测 |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Full Name | single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| Synonyms | UNG3, Single-strand selective monofunctional uracil DNA glycosylase, HMUDG, FDG, EC:3.2.2.- |
| Calculated Molecular Weight | 177 aa, 20 kDa |
| Observed Molecular Weight | 30 kDa |
| GenBank Accession Number | BC000417 |
| Gene Symbol | SMUG1 |
| Gene ID (NCBI) | 23583 |
| RRID | AB_3671365 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | Q53HV7 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
UNG(Uracil-DNA glycosylase) removes uracil in DNA resulting from deamination of cytosine or replicative incorporation of dUMP instead of dTMP. Thus, UNG plays a role in suppressing GC-to-AT transition mutations.The UNG gene encodes 2 isoforms that are individually targeted to the mitochondria and the nucleus(PMID:12369930).Defects in UNG are a cause of immunodeficiency with hyper-IgM type 5 (HIGM5).
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for SMUG1 antibody 83771-5-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |