Publication Spotlight
Celebrating the success of scientists citing Proteintech antibodies.
2022
February featured papers
|
“Finding SARS-CoV-2 vulnerabilities” Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and vulnerabilities.Kruse et al. Nature Communications. Products cited: Myc-Trap® Agarose, GFP-Trap® Agarose, GFP-Booster ATTO488 |
 |
|
|
“New role for ATG9A in fatty acid metabolism” The autophagy protein ATG9A enables lipid mobilization from lipid droplets.Mailler et al. Nature Communications. Products cited: GFP-Trap® Magnetic Agarose, TIP47 Rabbit PolyAb, ATG2A Rabbit PolyAb, ATG2B Rabbit PolyAb |
|
“MeCP2: the great protector” MeCP2 is a microsatellite binding protein that protects CA repeats from nucleosome invasion.Ibrahim et al. Science. Antibodies cited: MECP2 Rabbit PolyAb |
 |
|
|
“New modelling strategies for rare kidney diseases” A tissue-bioengineering strategy for modeling rare human kidney diseases in vivo.Hernandez et al. Nature Communications Antibodies cited: c-MYC Rabbit PolyAb, Human ENO2 ELISA Kit, PAX8 Rabbit PolyAb |
January featured papers
|
“Stopping the nucleosome invasion” Ibrahim et al., 2021. Science Cited Proteintech’s products: MECP2 rabbit polyclonal antibody |
 |
 |
“Balancing the control of nuclear shape” Wang et al., 2021. Cell Research Cited Proteintech’s products: KATNA1 Rabbit PolyAb, Septin 2 Mouse McAb, MAP1S Rabbit PolyAb, ARL13B Rabbit PolyAb |
|
“New models for kidney disease” Hernandez et al., 2021. Nature Communications Cited Proteintech’s products: Human ENO2 ELISA Kit, PAX8 Rabbit PolyAb, SIX2 Rabbit PolyAb. |
 |
 |
“A wasp-like conductor” Vila et al. 2021., Cell Metabolism Cited Proteintech’s products: FADS1 Rabbit PolyAb, GAPDH Mouse MonoAb |
2021 Publication roundup!
Thank you for the opportunity to support your work in 2021.
“Over the years we have witnessed countless discoveries and we couldn’t be prouder of everything our customers have achieved. We want to thank all of our customer for their tireless efforts and sacrifices to advance scientific discovery,”
- Dr. Jason Li, CEO
2021 Publication Roundup
Ground-breaking research published in top journals including:
Top research areas Neuroscience and Cancer |
 |
Top cited antibodies in 2021:
| GAPDH monoclonal (60004-1-Ig) |
BAX polyclonal (50599-2-Ig) |
| GAPDH polyclonal (10494-1-AP) |
BCL2 polyclonal (12789-1-AP) |
| Beta actin monoclonal (66009-1-Ig/60008-1-Ig) |
Caspase 3/p17/p19 polyclonal (19677-1-AP) |
| Beta actin polyclonal (20536-1-AP) |
E-cadherin polyclonal (20874-1-AP) |
October featured papers
September featured papers
Publication Spotlight highlights the new and exciting discoveries citing Proteintech antibodies published in scientific journals across the globe.
100,000 success stories and counting. What matters to Proteintech is the success of its antibodies in the hands of scientists - the people who make the discoveries that change and explain the world around us. In reaching 100,000 citations of Proteintech antibodies, our thanks go out to the talented researchers who made this milestone possible by using Proteintech antibodies in their work.
August featured paper
Cousin et al. Nature Genetics
‘Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome’
Ubiquitously expressed polypeptides that bind membrane lipids are known as spectrins; which bind ankyrins to line the cellular plasma membrane. The Spectrin meshwork is formed by heterodimeric α-spectrin and β-spectrin units. Mammalian neurons contain a wide range of spectrins including αII-spectrin and βI-V spectrins. Spectrins with ankyrins position and stabilise cellular adhesion molecules, membrane transporters, ion channels and scaffolding proteins. Previously, autosomal dominant variants in βIII-spectrin (SPTBN2) were identified and associated with late-onset spinocerebellar ataxia type 5, while autosomal recessive variants were found to be linked with childhood ataxia and intellectual disability. In the recent publication (PMID:34211179) Cousin et al. identified heterozygous variants of SPTBN1 affecting βII-spectrin stability, contributing to the neurodevelopmental disorder development.
The authors performed whole-exome sequencing (WES) on a cohort of 29 individuals from 28 different families with heterozygous SPRBN1 variants. WES identified 28 unique variants, of which: 22 were missense, three nonsense and three canonical splice-site variants. SPTBN1 variants were found to contribute to various neurological and behavioural changes causing neurodevelopmental syndrome. Seventeen unique SPTBN1 variants were classified as pathogenic, nine as likely pathogenic, and the two remaining of uncertain significance. Subsequently, a subset of SPTBN1 variants was introduced in GFP-targeted human βII-spectrin to assess the pathogenic mechanism of SPTBN1 variants. This experiment revealed that SPTBN1 variants in βII-spectrin cause changes to cytoskeleton structure and dynamics altering cellular morphology, and in turn causing disruptions in neuronal architecture which contribute to SPTBN1-associated syndrome. In turn, the effects of βII-spectrin haploinsufficiency and deficiency were assessed in vivo. βII-spectrin knockout (KO) mice showed that βII-spectrin loss of function can reduce neural connectivity, affecting global development and behaviour, and contributing to development of social behaviour impairments and autistic features which were observed with lesser severity in haploinsufficient animals.
This study reported de novo SPTBN1 variants that affect βII-spectrin stability through disruption of cytoskeleton organisation and dynamics, together with disrupted binding of key molecular partners. Collectively, these results revealed that SPTBN1 variants are linked to the development of neurodevelopmental disorder, intellectual disability, and behavioural comorbidities.
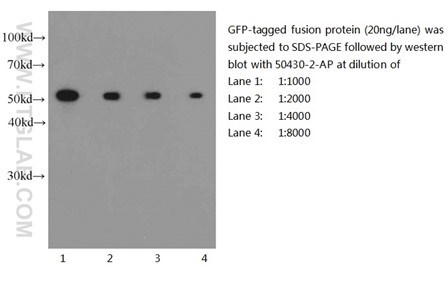
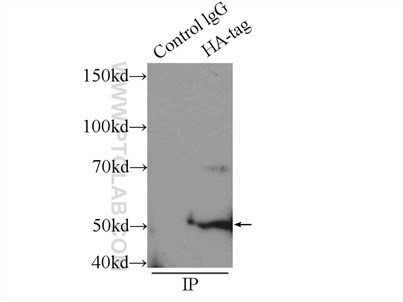
Six Proteintech antibodies were used in this study for Western Blot and Immunoprecipitation: GFP tag Monoclonal (66002-1-Ig), GFP tag Polyclonal (50430-2-AP), HA tag Polyclonal (51064-2-AP), Alpha-tubulin Monoclonal (66031-1-Ig) and 6xHis tag Monoclonal (66005-1-Ig).
 |
 |
|
Western blot of eGFP-tagged fusion protein with GFP tag Polyclonal antibody at various dilutions |
Immunoprecipitation experiment of transfected HEK-293 using HA tag Polyclonal antibody |
About the corresponding authors: Dr Margot A. Cousin is a Research Associate at Mayo Clinic in Rochester, USA; Dr Damaris N. Lorenzo is an Assistant Professor at the University of North Carolina where she runs a laboratory which studies ankyrins, spectrins and their partners in the aspects of cellular homeostasis and human disease.
July featured papers
June featured papers
|
“Heart failure has met it’s MARK(4)” MARK4 controls ischaemic heart failure through microtubule detyrosination.Yu et al. Nature Cited Antibodies: TTL Polyclonal antibody |
 |
 |
“SARS-CoV-2 has a nose for cilia” Nasal ciliated cells are primary targets for SARS-CoV-2 replication in early stage of COVID-19Ahn et al. Nature Cited Antibodies: Acetylated Tubulin(Lys40) Monoclonal antibody |
|
“Stress collides with ribosomes to activate cGAS” Translation stress and collided ribosomes are co-activators of cGASWan et al. Molecular Cell Cited Antibodies: ASCC1 Polyclonal antibody, IFIT2 Polyclonal antibody, GFP-Trap Magnetic Agarose |
 |
 |
“Metformin attenuates pulmonary inflammation by SARS-CoV-2” Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammationXian et al. Immunity Cited Antibodies: NFATC3 Polyclonal antibody, NDUFS4 Polyclonal antibody |
February featured papers
 |
“Nice to METTL3 you” Xu et al. Nature |
|
“Mitochondrial stabilizers” Wang et al. Cell Metabolism |
 |
 |
“Autophagy cargo selector” Zellner et al. Molecular Cell |
|
“Plant perception” Rhodes et al. Nature Communications Cited Products: ChromoTek RFP-Trap® Agarose, ChromoTek GFP-Trap® Agarose |
 |
January featured papers
|
“RNA handcuffs devious retroviruses” Chekmicki et al. Nature |
 |
 |
“Flava Flavirus” You know the time!” Heinrich Hoffman et al. Cell. Cited antibodies: TMEM41A |
|
“Like father, like son” Gordon et al. Science Cited Antibodies: TOM20 |
 |
 |
“Nowhere to run, nowhere to hide” Roth et al. Cell Chemical Biology |
December featured papers
 |
“Closed for a private TDP43 event”HSP70 chaperones RNA-free TDP43 into anisotropic intranuclear liquid spherical shells Yu et al. Science. Cited antibodies: TDP-43 (C-Terminal) |
“Metastatic information highway”A metastasis map of human cancer cell lines. Jin et al. Nature. Cited antibodies: SREBF1 |
 |
 |
“Viral prison break”Wang et al. Immunity Cited antibodies: GSTM1, MAVS, HRP-6*His-Tag, Beta Actin. |
“Traffic cops of the golgi”Cerrato et al. Cell Death and Differentiation Cited antibodies: TGN46 |
 |
November featured papers
“Pesky persistence to BRAF/MEK inhibitors”Melanoma Persister Cells Are Tolerant to BRAF/MEK Inhibitors via ACOX1-Mediated Fatty Acid Oxidation Shen et al. Cell Reports |
 |
 |
“Giving the DNA surgeon some room to operate”Gao et al. Nature Communications |
“Super Genomics finds pancreatic cancer Kryptonite”Biancur et al. Cell Metabolism Cited antibodies: FDFT1 |
 |
 |
“Not included in Miss Manners etiquette book”Daniloski et al. Cell |
October featured papers
“Cytomegalomaniac”Cytoplasmic control of intranuclear polarity by human cytomegalovirus Procter et al. Nature Cited antibodies: Emerin |
 |
 |
“Does this work for 2020 too?"A Cellular Mechanism to Detect and Alleviate Reductive Stress Manford et al. Cell Cited antibodies: FEM1B |
“You scratch my back and I’ll scratch yours”Di Marco et al. Cell Reports Cited antibodies: VEGF-A |
 |
 |
“The most cited paper by tooth fairies”Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth Krivanek et al. Nature Communications Cited antibodies: Smooth muscle actin |
September featured papers
“Protein buddy system”Structural basis for dimerization quality control. Mena et al. Nature Cited antibodies: PDCD6 |
 |
 |
“Schwann cells to the rescue”A glycolytic shift in Schwann cells supports injured axons Babetto et al. Nature Neuroscience Cited antibodies: RICTOR, Raptor, OGDH, IDH3A, Citrate synthase, LDHB, PGK1, PFKM, GPI |
“Cancer slugger”Glutamine depletion regulates Slug to promote EMT and metastasis in pancreatic cancer. Recouvreux et al. J Exp Med |
 |
 |
“VAMPing up ER-phagy”CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. Nthiga et al. EMBO J |
August featured papers
Download the printer friendly version here
“It’s NRF2 or Nothin”Activation of NRF2 ameliorates oxidative stress and cystogenesis in autosomal kidney disease. Lu et al. Sci Trans Med |
 |
 |
GALC it upWeinstock et al. Neuron |
Sympathy for the adipose tissueA leptin-BDNF pathway regulating sympatheticinnervation of adipose tissue Wang et al. Nature Cited antibodies: UCHL1 |
 |
 |
MegaDalton Protein to the rescueNaked mole-rat very-high-molecular mass hyaluronan exhibits superior cytoprotective properties. Takasugi et al. Nat. Comm |
The Pearly CiliaTALPID3 and ANKRD26 selectively orchestrate FBF1 localization and cilia gating Gates Yan et al. Nat. Comm Cited antibodies: ARL13B , FBF1 , GAPDH , IFT140 , KIAA0586, NKIRAS2, SCLT1, TCTN1
|
 |
Lu et al.
Science Translational Medicine
Activation of NRF2 ameliorates oxidative stress and cystogenesis in autosomal dominant polycystic kidney disease
Autosomal dominant polycystic kidney disease (ADPKD) is a deadly genetic disorder caused by mutations in PKD1/2 genes. It is characterized by renal cysts that fill with fluid, leading to kidney failure. Current therapeutic approaches provide only a few years of life after diagnosis, with kidney transplants being the most effective current treatment. In a recent study (PMID: 32727915), Yi Lu and colleagues discover a potential pharmaceutical target for this disease.
To find therapeutic targets for ADPKD, Lu and colleagues compared the proteomic signatures of kidneys from genetic ADPKD model (Pkd1-/-) and wild-type mice, and found that proteins involved in mitochondrial function were downregulated in ADPKD kidneys. Investigating this further, it was discovered that reactive oxidative species (ROS) levels were high in these tissues and that increasing ROS levels correlated with disease progression in patients. Consequently, they examined whether a key transcription factor in ROS maintenance, NRF2, may be responsible. Not only were NRF2 levels lower in ADPKD kidneys, but all its gene targets were also decreased. Knocking this gene out in a genetic ADPKD mouse model (Pkd1−/−;Nrf2−/−) increased disease severity. Consistent with NRF2 playing a protective role, increasing NRF2 activity by pharmacological inhibition of its two negative regulators improved the health of ADPKD mice. To understand the mechanism, the authors next examined the genomic effects of inducing NRF2 activity in the ADPKD 9-12 cell line. Using ChIP-SEQ, they found that NRF2 was bound to enhancers to the active histone markers H3K4Me1 and H3K27ac, suggesting increased target gene transcription. Comparing ChIP-SEQ data of ADPKD cells with and without pharmacological downregulation of NRF2 inhibitors, it was revealed that histone acetyltransferase P300 has a similar footprint to NRF2, suggesting NRF2 recruits P300 to add activating markers to its enhancers. Further supporting this, the enhancer targets all had increased activity in the treatment condition. Diving deeper into the enhancer mechanism, the authors tested the possibility that NRF2 forms a phase-separated complex with MED1 of the transcription machinery to promote transcription. Using an in vitro droplet formation assay, they showed that NRF2 can form phase-separated compartments and that MED1 can be included in these compartments. Consistent with this hypothesis, ChIP-SEQ revealed that NRF2 and MED1 occupy the same genomic regions. Overall, the group’s work suggests that loss of NRF2 is critical to the progression of ADPKD and that targeting its regulators may be an effective therapeutic strategy.
Three Proteintech antibodies were used in this study for Western blot: KEAP1 (10503-2-AP), NQO1 (11451-1-AP), and alpha-tubulin (11224-1-AP).
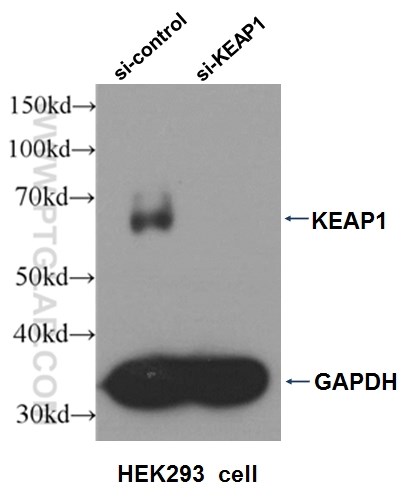
Western blot analysis of HEK293 cells that were untransfected (Lane 1) and transfected with si-KEAP1 vector (Lane 2) using KEAP1 antibody (10503-2-AP).
About the corresponding author, Dr. Yupeng Chen: Yupeng Chen is a professor in the Collaborative Innovation Center of Tianjin for Medical Epigenetics at Tianjin Medical University in China. His laboratory focuses on kidney diseases and epigenetics.
Origins and Proliferative States of Human Oligodendrocyte Precursor Cells
Much of human evolution can be attributed to the increase in size and complexity of the cerebral cortex. Cerebral white matter is developed from myelinating oligodendrocytes, arising from the differentiation of oligodendrocyte precursor cells (OPCs). A key challenge in this field is to understand their development in humans, as much previous work was performed in rodents, which have vastly decreased white matter. Humans make use of an enlarged cortical germinal zone, the outer subventricular zone (OSVZ), which is populated with outer radial glia (oRG) and progenitors for OPCs. How human OPC production is developed from this zone is poorly understood, and understanding this process is key to finding new treatments to traumatic brain injuries in addition to deepening our understanding of white matter expansion.
In this work (PMID: 32679030), Huang and colleagues studied the transcriptional profile of embryonic human brain tissue samples using unbiased capture in combination with immunopanning to identify high quality cells with classic OPC markers. Their analysis revealed a subset of cells containing markers for both neural progenitor cell (NPC) and OPC lineage, suggesting a transitional pre-OPC stage. They found over 10% of OPCs express low levels of transcriptional markers for both OPCs and NPCs, which, along with their immature morphology, suggested the role of these cells is a transitional intermediate between OPCs and NPCs. They then tested whether potential pre-OPC cells were capable of becoming OPCs in vitro. After 7 days in culture, around half of the cells demonstrated a reduction in progenitor markers and an upregulation in mature OPC markers such as OLIG2 and PDGFRA, indicating that these progenitors can generate OPCs. They then investigated the origin of the pre-OPC cells. One candidate was neurogenic oRGs, which play a role in gray matter expansion in humans, because the transcriptional profile of the pre-OPC cluster also contained oRG gene markers, such as HOPX. Furthermore, immunohistochemical analysis revealed that a small percentage of oRGs and pre-OPCs in the OSVZ both expressed HOPX. To test whether HOPX-expressing oRGs could generate OPCs, these cells were isolated and cultured in vitro. By day 10, roughly 17% of cells expressed mature OPC markers (OLIG2 and PDGFRA), suggesting that oRGs can produce OPCs and thus may be an additional source of OPCs in the developing human brain. Time-lapse imaging and GFP labeling showed that pre-OPCs had mitotic somal translocation during division, similar to oRGs, and regrew processes similar to OPCs. The authors also observed that OPCs divided symmetrically and frequently, in comparison to mouse embryonic OPCs, which divide only once in 5 days. These findings indicate the properties, derivation, and proliferation of OPCs from oRGs in the developing human brain, an important part of white matter expansion.
Proteintech’s HOPX antibody (11419-1-AP) was used in this study as a marker of oRG cells, utilizing immunofluorescence to track the development of oRGs to pre-OPCs in vitro.
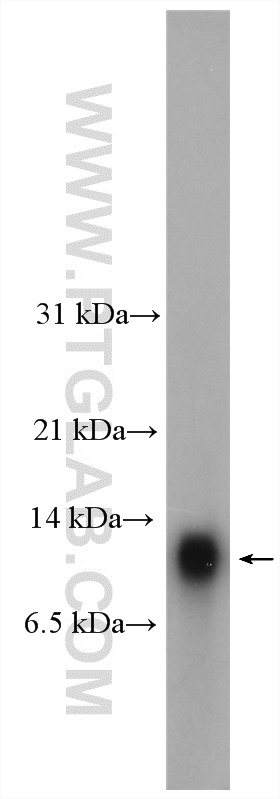
Mouse lung tissue were subjected to SDS PAGE followed by western blot with 11419-1-AP (HOPX antibody) at dilution of 1:300 incubated at room temperature for 1.5 hours.
About the corresponding author: Arnold Kriegstein is a Professor of Neurology at the University of California, San Francisco. His work focuses on the embryonic development of neural cells.
Reversing a model of Parkinson’s disease with in situ converted nigral neurons
Parkinson’s disease is a neurodegenerative disorder affecting motor function. The major neurons affected are dopaminergic neurons in the substantia nigra pars compacta in the midbrain. A potential therapeutic approach for this disease is regenerative medicine. Previous work had demonstrated that repressing transcription factors PTB and nPTB can convert human fibroblasts to neurons. In this work (PMID: 32581380), Qian et al. extend this insight to astrocytes, demonstrating that targeting a single protein – PTB – can efficiently convert astrocytes to dopaminergic neurons. Remarkably, these converted neurons restored motor function in a Parkinson’s mouse model, paving the way for in vivo regenerative medicine.
Due to the plasticity of astrocytes, Qian and colleagues hypothesized that astrocytes, like fibroblasts, could be transcriptionally reprogrammed into neurons. Comparing the transcriptional programming between neurons, astrocytes, and fibroblasts, they found that the necessary nPTB repression loop was already in place for astrocytes and that only the PTB repression loop needed to be implemented to facilitate conversion. To test whether PTB knockdown was sufficient for neural conversion, Qian and colleagues delivered a lentiviral shRNA construct to midbrain astrocytes in mice. The morphology, marker expression, and function of the converted astrocytes were consistent with dopaminergic neurons in the targeted region, thus validating their conversion. Additionally, the axons of the converted astrocytes targeted the appropriate brain subregions. Qian and colleagues then tested the ability of converted astrocytes to restore function in a Parkinson’s mouse model. Lentiviral delivery of PTB shRNA resulted in a partial restoration of the number of dopaminergic neurons. Furthermore, dopamine biogenesis and activity-induced release of the converted astrocytes were nearly similar to controls, indicating their potential to sufficiently replace lost neurons. To test the functionality of these converted neurons in vivo, mice were treated with PTB shRNA and they performed at a similar level as control mice in motor-skills tests. Lastly, using a chemogenetic approach targeting dopaminergic neurons, they showed that the converted astrocytes were responsible for the effects. Since lentiviral delivery is not clinically viable, Qian and colleagues then tested whether injection of a more practical therapy, anti-sense oligos, could achieve the same results. Marker expression and behavioral testing suggested that this approach has similar efficacy to lentiviral delivery. Though proof-of-concept, these results suggest an elegant genetic method for limiting the effects of Parkinson’s disease, and could revolutionize the treatment of this disease moving forward.
9 Proteintech antibodies were used in this study. VMAT2 (20873-1-AP), DAT (22524-1-AP), ALDH1L1 (17390-1-AP), Calbindin D28K (14479-1-AP), OTX2 (13497-1-AP), SOX6 (14010-1-AP), and S100b (15146-1-AP) were used as dopaminergic neuron markers. Cux1 (11733-1-AP) was used as a cortical neuron marker and beta-actin (66009-1-IG) was used as a loading control.
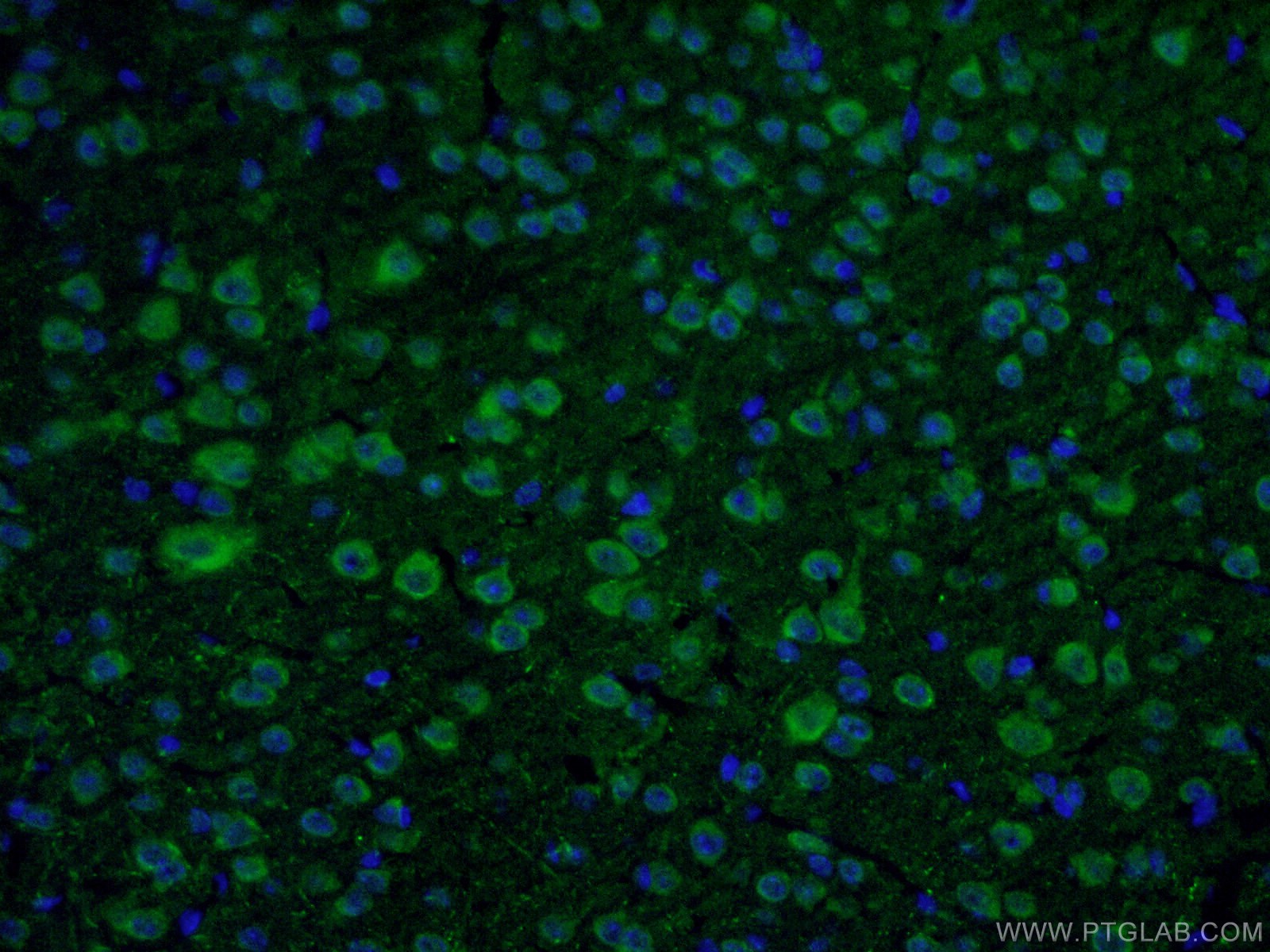
Immunofluorescent analysis of fixed mouse brain tissue using 22524-1-AP (1:50, green) and DAPI (blue). About the corresponding author, Xiang-Dong Fu, PhD: Dr. Fu is a Distinguished Professor of Cellular and Molecular Medicine at the University of California, San Diego. His lab focuses on RNA regulation and fate determination.
Activation of GCN2/ATF4 signals in amygdala PKC-δ neurons promotes WAT browning under leucine deprivation
Recently, the conversion of white adipose tissue (WAT) into a high energy consuming state similar to brown adipose tissue has received increased attention due to its therapeutic potential to combat obesity and metabolic disorders. WAT browning increases the metabolic capacity of WAT cells by promoting the use of free fatty acids as fuel, occurring in response to prolonged cold exposure, intermittent fasting, and low-protein/high-carbohydrate diets. Specifically, branched chain amino acids such as leucine and valine are highly important in metabolic pathways regulating lipid metabolism and glucose utilization. Distinct neurons in the hypothalamus in the brain, a key area for processing metabolic signals, are primarily responsible for the central control of processes that influence WAT browning. Recently, certain neurons in the amygdala have been shown to influence certain metabolic processes, an area not traditionally known for metabolic functions.
Yuan and colleagues published a recent paper in Nature Communications, describing the role of leucine deprivation in the regulation of WAT browning. They discovered that dietary leucine deprivation induced WAT browning, but that this induction can be blocked by inhibiting protein kinase c-δ (PKC-δ) neurons in the amygdala, indicating the high-level involvement of these neurons in this process. The authors went on to describe a possible mechanism for this phenomenon through general control nonderepressible 2 (GCN2), a neuronal amino acid sensor in the amygdala, and its downstream effector, activating transcription factor 4 (ATF4). Specifically, deleting GCN2 from PKC-δ amygdala neurons reduced the amount of WAT browning seen following leucine deprivation. Supporting this finding, they showed that blocking activation of ATF4 in these neurons was also able to decrease markers of WAT browning. These results highlight a novel role of central amino acid sensing through GCN2 and its interactions with ATF4 in the amygdala, shedding light on possible dietary interventions to induce WAT browning and thus providing novel pathways to help treat metabolic disease.
Proteintech’s ATF4 antibody (10835-1-AP) was used in this study to confirm ATF4 knockdown in PKC-δ in amygdala neurons. This antibody has been cited 125 times and validated in WB, IP, IHC, IF, FC, and ELISA.
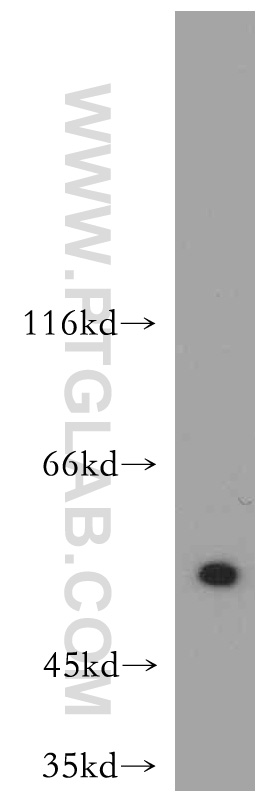
A431 cells were subjected to SDS PAGE followed by western blot with 10835-1-AP at a dilution of 1:500 and incubated at room temperature.
About the corresponding author:
Feifan Guo. Professor Guo is a professor at the Shanghai Institute of Nutrition and Health, and her work focuses on the molecular mechanisms underlying nutrition-related metabolic diseases.
Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate
Obesity is a growing problem across the world due, in part, to increased fructose consumption. It is well known that fructose causes the liver to produce lipids in a process called de novo lipogenesis (DNL), so a key enzyme in converting fructose to acetyl-CoA, ACLY, has been an attractive therapeutic target. However, in a recent Nature paper (PMID: 32214246), Zhao and colleagues demonstrate that ACLY is not involved in fructose-induced DNL and that microbiota metabolize dietary fructose into acetate to fuel DNL in the liver.
To investigate the role of ACLY in hepatic DNL, Zhao and colleagues assayed the response of ACLY KO mice to high fructose consumption compared to wild-type controls. Surprisingly, both KO and control mice showed the same DNL response, suggesting that this process does not require ACLY. To find clues to this alternative route to DNL, they investigated the transcriptional response to fructose. They found that ACSS2, which converts acetate to acetyl-CoA, significantly increases upon fructose consumption. This result raised the possibility that acetate may be playing a role. Consistent with acetate being the favored substrate for fructose-induced DNL, hepatocytes incorporate acetate more readily than fructose in DNL. In vivo, DNL intermediates appear much later than fructolytic intermediates and increase significantly after acetate appears in the portal blood. A possible source of acetate is the gut microbiota. To test this, Zhao and colleagues performed an oral gavage of radioactive-labeled fructose to wild-type mice with and without antibiotic treatment. Though fructolytic intermediates were unchanged between the two groups, antibiotic-treated mice had a lower DNL response. Furthermore, an oral gavage of fructose and radioactive-labeled acetate resulted in similar DNL responses between groups. Interestingly, the transcriptional response to fructose was independent of antibiotic treatment. Overall, Zhao et al.’s study suggests an interplay between the microbiota and organs in DNL, and casts doubt on the potential therapeutic efficacy of ACLY inhibitors for obesity and non-alcoholic fatty liver disease.
Proteintech’s ACLY antibody (15421-1-AP) was used in this study to confirm their knockout mouse model and show ACLY induction after fructose consumption. This antibody has been validated in WB, IP, IHC, IF, and FC.
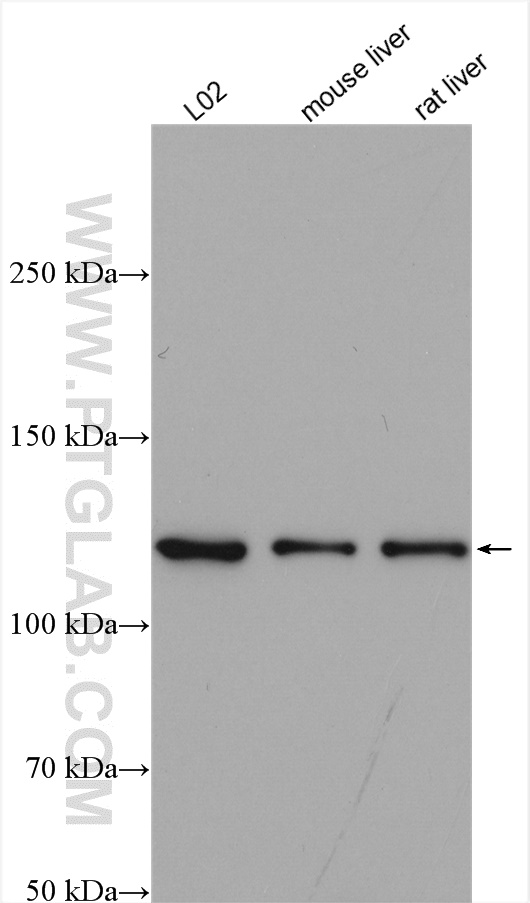
Various lysates were subjected to SDS-PAGE followed by Western blot with ACLY antibody (15421-1-AP) at 1:2000 dilution and incubated at room temperature for 1.5 hours.
About the corresponding author, Kathryn E. Wellen:
Dr. Wellen is an associate professor of Cancer Biology at the University of Pennsylvania Perelman School of Medicine. Her research focus is the biochemistry of cancer and metabolic disease.
Tropism, Replication Competence, and Innate Immune Responses of the Coronavirus SARS-CoV-2 in Human Respiratory Tract and Conjunctiva: An Analysis in Ex-Vivo and In-Vitro Cultures
A key part of mitigating the risk of infection by the novel coronavirus is understanding how it enters the body and which parts of each organ it infects. In a recent Lancet Respiratory Medicine article (PMID: 32386571), Hui and colleagues explored the ability of SARS-CoV-2 to infect different tissues.
To investigate organ tropism of SARS-CoV-2, Hui et al. first generated ex vivo cultures of bronchus, ocular conjunctiva, and lung tissues. They then infected them with either SARS-CoV-2, SARS-CoV, H1N1, or MERS-CoV and assessed infection after 4 days using immunohistochemical staining of viral components. All the tissues showed viral staining, indicating infection. Next, Hui and colleagues compared the replication rate of the different viruses in their ex vivo cultures. In comparison to MERS-CoV, SARS-CoV-2 replication was nearly identical in the bronchus, and less than MERS-CoV in the lung. In comparison to SARS-CoV, SARS-CoV-2 replication was higher in the bronchus and similar in the lung and ocular conjunctiva. Interestingly, proinflammatory cytokines were at lower levels after infection with SARS-CoV-2 than H1N1, MERS-CoV, and SARS-CoV. Overall, the study suggests that the bronchus, lung, and ocular conjunctiva are viable entries for SARS-CoV-2 infection and this may help inform health policies.
Proteintech’s CC10 antibody (10490-1-AP) was used to label club cells in their bronchus ex vivo cultures. This antibody has been validated for use in WB, IHC, and IF.
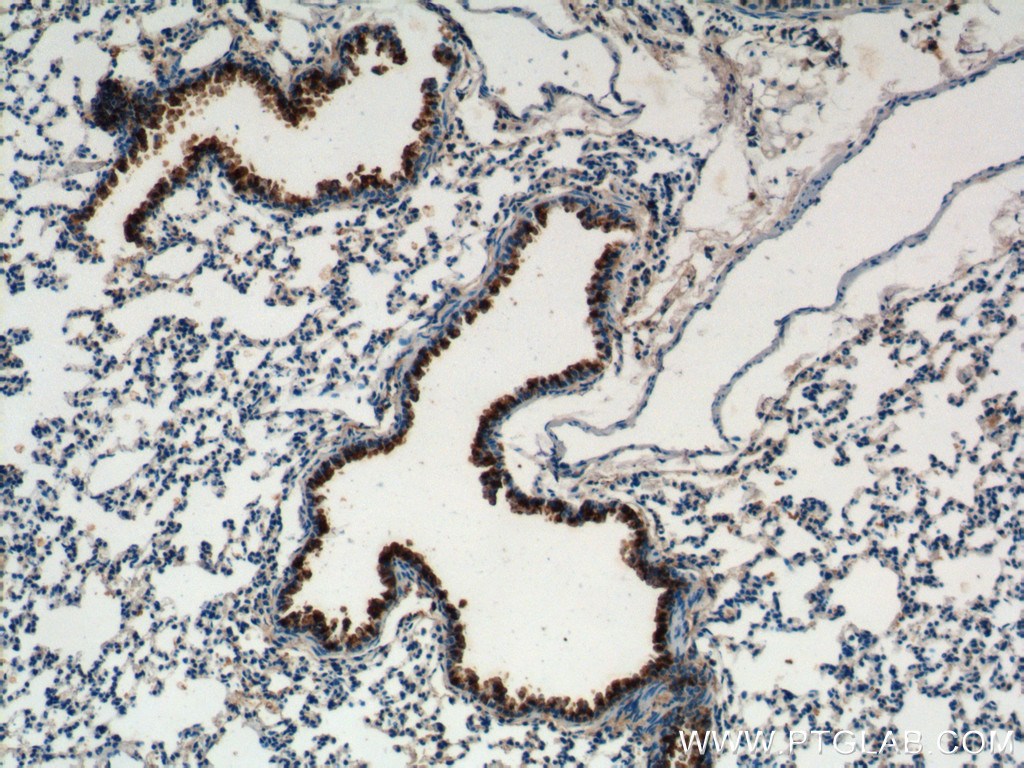
Immunohistochemical analysis of mouse lung tissue using CC10 antibody (10490-1-AP, 1:200).
About the corresponding author, Michael Chan:
Michael Chan, PhD is an associate professor in the School of Public Health at the University of Hong Kong. His research focus is on coronavirus and influenza pathogenesis.
5/15/20
Proline biosynthesis is a vent for TGFB-induced mitochondrial redox stress
Underneath a wound as minor as a simple paper cut is a complex coordinated response of signaling and protein production. One key growth factor in wound healing is TGFB. This signal causes fibroblasts to generate collagens, essential extracellular matrix components. Collagens are enormous proteins rich in proline and glycine, so TGFB signaling needs to reprogram metabolism to enable production. In a recent EMBO paper (PMID: 32134147), Schworer et al. show that TGFB signaling enables fibroblasts for collagen production by rapidly increasing the rate of oxidative phosphorylation, setting off a cascade of events. The high activity of the TCA cycle creates excessive redox potential, which in turn promotes proline biosynthesis. The accumulation of this amino acid then enables the production of proline-rich collagens.
To understand the metabolic shift during wound healing, Schworer and colleagues first examined the metabolic state of NIH-3T3 fibroblasts stimulated with TGFB. Measuring ATP and metabolite levels, they found that TGFB increases glutamine consumption, protein translation, TCA cycle activity, and oxidative phosphorylation. Consistent with this growth factor stimulating collagen deposition, TGFB-conditioned fibroblasts elevated synthesis of collagen’s major amino acids: proline and glycine. The link between TGFB and proline levels is not well-characterized, so Schworer and colleagues investigated whether proline biosynthesis – a process that consumes NADH and NADPH – is a redox vent from elevated TCA cycle activity during TGFB stimulation. To test this, they ectopically expressed NADH and NADPH oxidases to act as competing redox sinks and found that these conditions eliminated proline biosynthesis. Consistent with their hypothesis, disabling proline biosynthesis by genetically deleting key enzymes resulted in ROS accumulation. Schworer et al.’s work suggests a novel link between signal transduction and metabolism to increase collagen production in fibroblasts and may provide insight into fibrotic diseases.
Four Proteintech antibodies were used in this study. SLC25A1 antibody (15235-1-AP) was used to verify gene knockout in an experiment assessing TGFB’s effect on ROS production. PYCR1 (20962-1-AP) and PYCR2 (17146-1-AP) antibodies were used to show that TGFB upregulates the proline biosynthetic pathway. Lastly, using the Collagen IV (55131-1-AP) antibody, the authors demonstrated TGFB’s potent effect on collagen IV deposition.
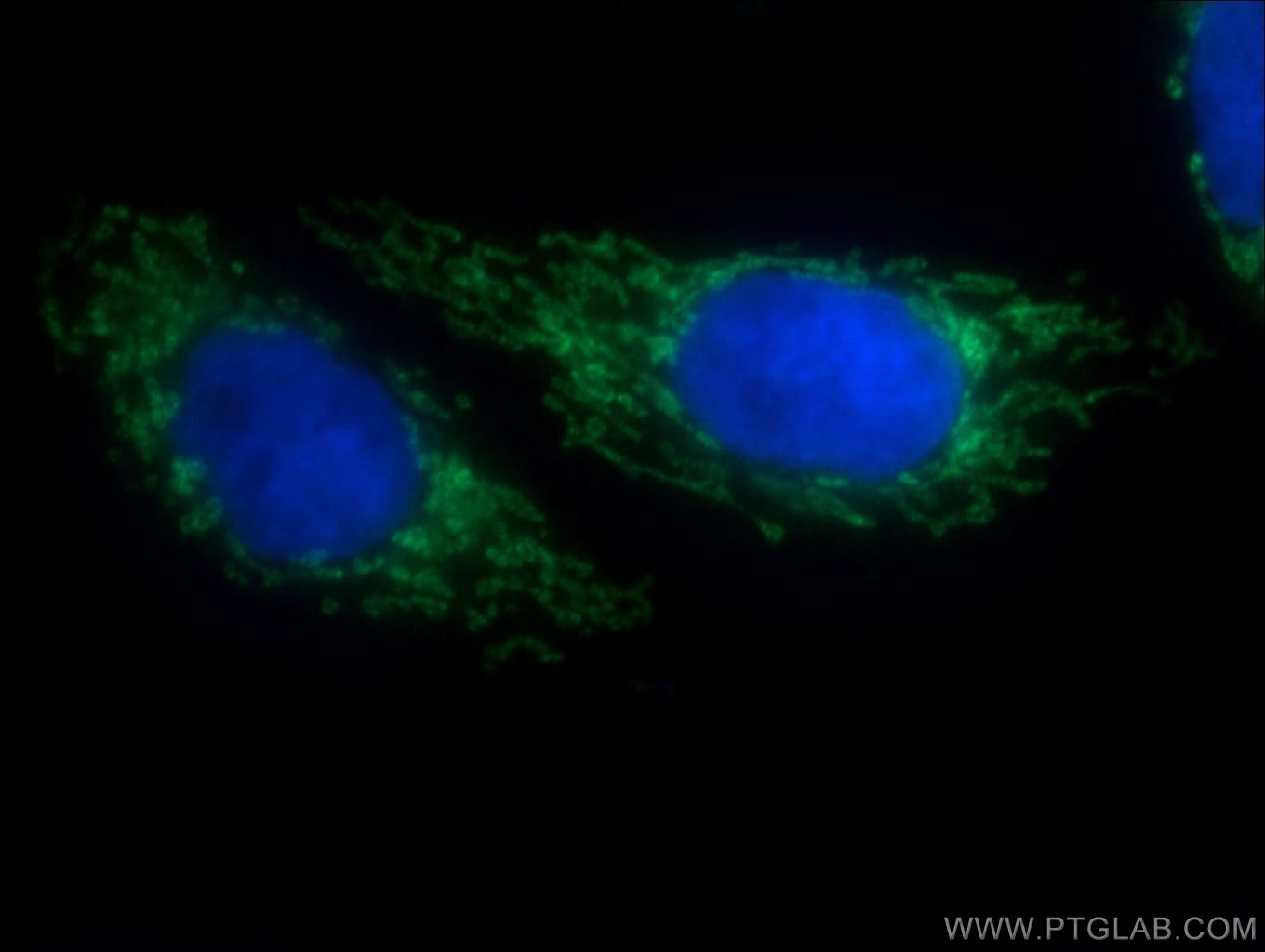
Immunofluorescent analysis of HepG2 cells using 15235-1-AP (SLC25A1 antibody, 1:50) and Alexa Fluor488-conjugated AffiniPure goat anti-rabbit IgG(H+L) with nuclear DNA stained by DAPI.
About the corresponding author, Craig Thompson:
Dr. Thompson is the current president of the Memorial Sloan Kettering Cancer Center. His work focuses on cancer metabolism and immunology. He has over 30 patents and has founded several biotechnology companies.
A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion
Like any bustling metropolis, the cell needs many on-ramps onto the highway to exit town. The major on-ramp for secreted proteins is through recognition of a specific amino acid motif by a signal recognition particle and translocation into the ER. However, some proteins lacking this motif are secreted, such as IL-1B, suggesting alternative routes of transport. In a recent Cell paper (PMID: 32272059), Zhang et al. uncover the mechanism behind the unconventional secretion of IL-1B.
To identify the machinery involved in vesicular translocation of IL-1B, Zhang and colleagues created a “trap” for protein channels. Since IL-1B’s vesicular translocation had previously been shown to be dependent on its own unfolding, they added a DHFR tag that can be maintained in the folded state in the presence of aminopterin. The idea is that the IL-1B-DHFR recombinant protein plus aminopterin would become trapped in the pore and mass spectrometry of the cross-linked complex would allow for the identification of the translocation machinery. Their assay revealed TMED10 as a potential component, which was validated using genetic manipulation, usage of different cell lines, and a mouse model. To understand the interaction between TMED10 and IL-1B, they performed a series of co-immunoprecipitation experiments with different TMED10 mutants. This analysis suggested that IL-1B directly interacts with TMED10’s C-terminal domain for translocation. They then looked at whether TMED10 is involved in the unconventional secretion of other proteins. Consistent with a broad role, genetic knockdown of TMED10 decreased secretion of many interleukin family members and other proteins. To understand the mechanism of TMED10-mediated translocation, Zhang and colleagues created an in vitro proteoliposome protection assay. Through this system, they determined that TMED10 requires unfolding of the cargo target and is sufficient for driving translocation. Moving back into a cellular system to view the events after translocation, they determined that TMED10-bearing vesicles interface with the ER-Golgi intermediate compartment using immunofluorescence and confirmed that these vesicles contain IL-1B using a split-GFP construct. Overall, their work suggests that TMED10 is an essential translocator for the unconventional secretion of IL-1B and other proteins.
Three Proteintech Antibodies were used in this study: GRP94 (14700-1-AP), TMED9 (21620-1-AP), and TMED10 (15199-1-AP). All were used for Western blotting, while the TMED10 antibody was used as the major antibody for determining TMED10 levels and localization throughout the study. The authors also validated it in KO cell lines (Figure 1 in PMID: 32272059).
Various lysates were subjected to SDS-PAGE followed by Western blotting with 15199-1-AP (TMED10 antibody, 1:2500).
About the corresponding author:
Dr. Liang Ge is an assistant professor at Tsinghua University in Beijing, China. His research focuses on understanding the mechanisms behind unconventional secretion and organelle interaction.
Myc-mediated SDHA acetylation triggers epigenetic regulation of gene expression and tumorigenesis
In a recent paper, Li and colleagues examined the role of transcription factor cMyc, deregulated in many cancers, on the epigenetic regulation of gene expression. Using mass spectroscopy, western blotting, and immunoprecipitation in an expression-inducible cell system, they discovered that cMyc is responsible for the lysine acetylation of many mitochondrial proteins, including succinate dehydrogenase complex subunit A (SDHA), an enzyme in the tricarboxylic (TCA) cycle. cMyc causes proteasome-dependent degradation of SIRT3 deacetylase, which leads to increased acetylation of SDHA at Lys335 by cMyc, which in turn decreases the enzymatic activity of SDH. As a consequence, increased levels of succinate lead to changes in gene expression and increased histone methylation (H3K4me3), as detected by ChIP-seq, due to lower activity of KDM5A demethylase. Using mouse xenograft experiments and analysis of human cancer slices, Li and colleagues demonstrated that the tumor progression of cancers with deregulated Myc is at least in part mediated by higher SDHA acetylation levels uncovering an important mechanism of the epigenetic regulation of cellular metabolism in cancer by Myc.
About the corresponding author:
Dr Ping Gao works at the University of Science and Technology of China in Hefei (China) and focuses on studying metabolism in cancer and stem cells with a particular interest in the regulation of metabolism by the cMyc oncogene.
| ACAT1 antibody | SDHA antibody |
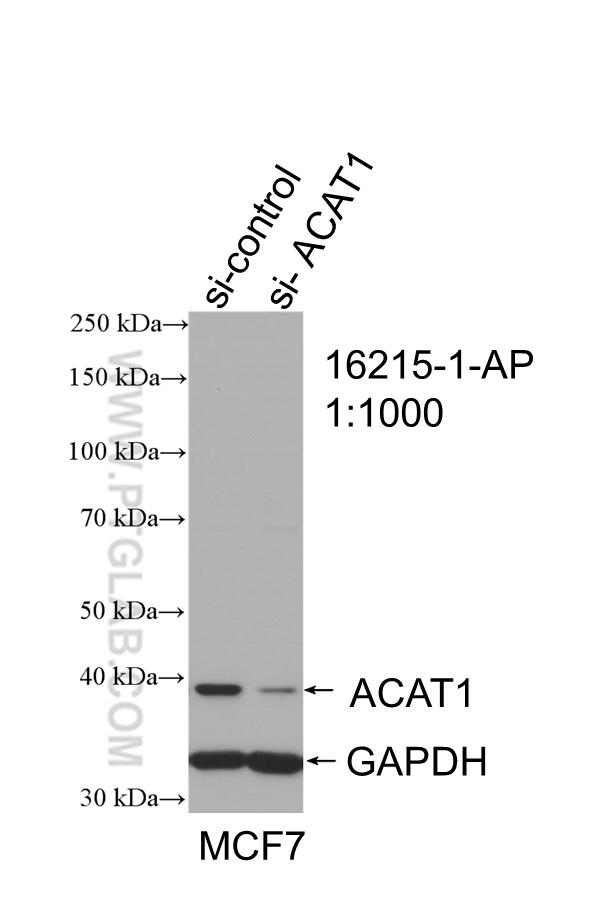 |
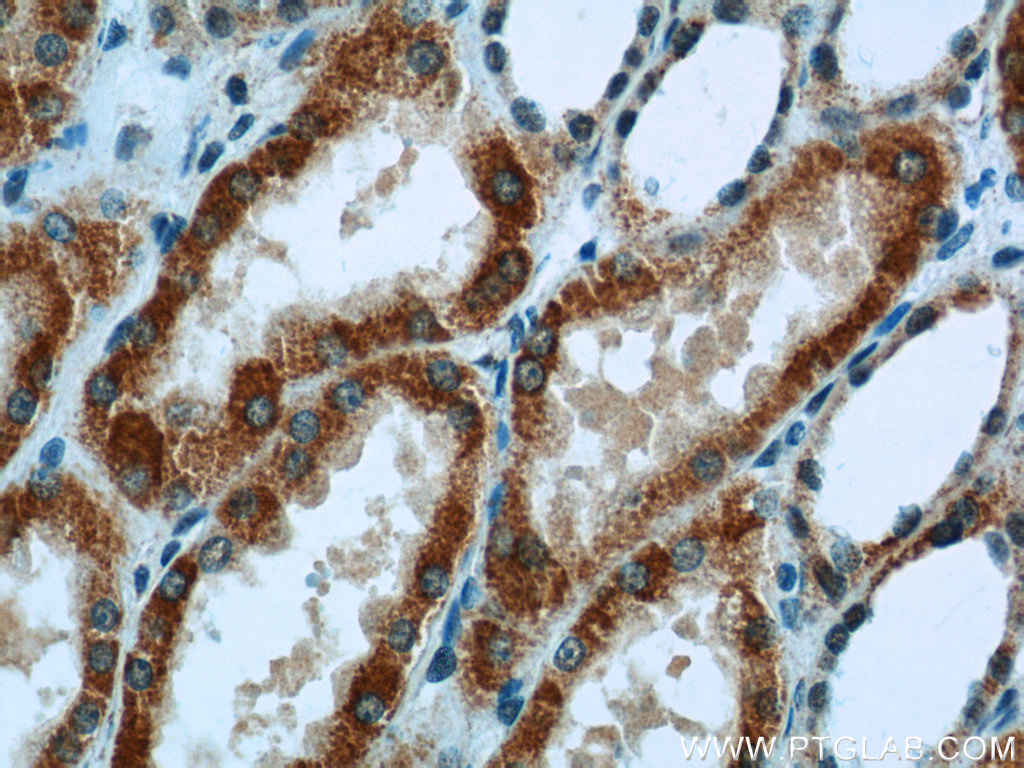 |
| WB result of ACAT1 antibody (16215-1-AP; 1:1000; incubated at room temperature for 1.5 hours) with sh-Control and sh-ACAT1 transfected MCF-7 cells | Immunohistochemical analysis of paraffin-embedded human kidney tissue slide using 14865-1-AP (SDHA Antibody) at a dilution of 1:50 (under 40x lens). |
A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol
Monitoring the stress of the mitochondria is critical to eukaryotic cell survival. How that stress is communicated to the rest of the cell and initiates the integrated stress response is unknown. Writing in Nature (PMID: 32132706), Fessler and colleagues suggest that mitochondrial stress triggers the cleavage and subsequent release of mitochondrial DELE1 into the cytosol where it binds to HRI to initiate the stress response.
To identify proteins involved in the communication of mitochondrial stress, Fessler et al. designed a genetic screen using haploid human HAP1 cells with a cell stress response reporter. They generated mutants using a gene trap approach, then subjected each clone to one of three chemical stresses (tunicamycin, CDDO, and CCCP) and assayed the effect on initiating the integrated stress response via their reporter. Using three separate treatments allowed them to filter out global regulators and identify mechanism-specific stress regulators. To find regulators of ionophore-induced mitochondrial stress, they focused on the signature of CCCP, a drug that disrupts oxidative phosphorylation and causes immense mitochondrial stress. Two proteins were enriched in their analysis: DELE1 and HRI. After confirming their roles in mediating the stress response using different cell lines and other methods of inducing mitochondrial stress, they investigated the relationship between DELE1 and HRI. Using a series of genetic experiments with DELE1 and HRI knockouts, they found that HRI expression can rescue induction of the stress response, but DELE1 expression cannot rescue HRI knockouts. Additionally, removing the DELE1 mitochondrial targeting sequence constitutively activated the stress response. These results raised the possibility that translocation of DELE1 conveys the message of mitochondrial stress to HRI. Consistent with this, DELE1 is cleaved and observed in the cytosol upon CCCP treatment. Additionally, immunoprecipitation and mutagenesis experiments revealed that cleaved DELE1 directly binds to HRI under stress. To link DELE1 to the initial mitochondrial stress, they investigated whether mitochondrial stress-induced protease OMA1 has an impact on this protein. They found that knocking out OMA1 abrogated cleavage of DELE1 under various types of mitochondrial stress. Fessler et al.’s experiments suggest a model where mitochondrial stress leads to OMA1 activation and cleavage of DELE1, which then translocates to the cytosol to activate HRI and induce the integrated stress response.
Proteintech’s HRI/eIF2AK1 (20499-1-AP) and OMA1 (17116-1-AP) antibodies were used in this study. Anti-HRI/eIF2AK1 was used in the co-IP experiment as the pull-down antibody to show that HRI and DELE1 are binding partners. Anti-OMA1 was used in WB to monitor activation upon mitochondrial stress.
Immunohistochemical analysis of paraffin-embedded human liver tissue slide using OMA1 antibody (17116-1-AP) at dilution of 1:200.
About the corresponding author:
Lucas T. Jae, Ph.D., is a professor at the Gene Center and Department of Biochemistry at Ludwig-Maximilians-Universität München in Germany. His work focuses on uncovering the genetics of disease and mitochondrial fidelity.
Pyridoxine induces glutathione synthesis via PKM2-mediated Nrf2 transactivation and confers neuroprotection
Parkinson’s disease (PD) is a debilitating neurodegenerative disorder with limited therapeutic options. A promising area of research is the role of damage from reactive oxidative species (ROS) in the pathogenesis of this disease.Oxidative damage caused by ROS in patients’ brains are thought to be one of the main underlying causes of PD. A major player in maintaining healthy levels of ROS is glutathione, a reducing agent.
To study the role of glutathione in PD pathogenesis, Wei and colleagues (PMID: 32071304) recently examined molecular causes of glutathione deficiency occurring in patients with PD. Using agonists and antagonists of dopamine receptors, they identified that activation of dopamine D2 receptor (DRD2) in astrocytes activates glutathione synthesis both in vitro and in vivo. RNA sequencing of astrocytes revealed that glutathione production in response to DRD2 stimulation is mediated by a transcription factor Nrf2. Pull-down studies using anti-Nrf2 antibody combined with mass spectrometry identify an important cofactor, PKM2, whose dimerization is mediated by β-arrestin-mediated D2D2 signaling. Dependence of glutathione levels in astrocytes on dopamine explains its lower levels in patients with PD. Supplementation with pyridoxine, which promotes DMD2 dimerization, resulted in higher glutathione levels in cultured astrocytes, as well as reduced dopaminergic neuron loss in the mouse model of PD.
Proteintech’s rabbit anti-NRF2 (16396-1-AP) was used in this study to identify its binding partners co-regulating glutathione synthesis. In chromatin immunoprecipitation assays they identified DNA sites associated with activated Nrf2 and in a proximity ligation assay to demonstrate interaction between Nrf2 and PKM2 in astrocytes nuclei.
Proteintech’s rabbit anti-GCLC (12601-1-AP) was used to examine effects on glutathione synthesis. GCLC is a rate-limiting enzyme in glutathione biosynthesis.
Proteintech’s rabbit anti-GCLM (14241-1-AP) was used to examine effects on glutathione synthesis. GCLM is a rate-limiting enzyme in glutathione biosynthesis.
About the corresponding author:
Gang Hu works at the Department of Pharmacology at universities in Nanjing and focuses on studying astrocyte biology, signaling, microRNA-mediated regulation of gene expression and Parkinson’s disease.
Increased Neural Progenitor Proliferation in a hiPSC Model of Autism Induces Replication Stress-Associated Genome Instability
Autism spectrum disorder (ASD) is a neurodevelopmental disorder affecting millions globally. How the growth and function of neurons are impacted during embryogenesis is a key question in the field.
Previously, studies have found a correlation between macroencephaly and autism, suggesting a link between increased proliferation and loss of neuronal function. In the cover story for Cell Stem Cell, Wang et al. suggest that an accumulation of double-strand breaks owing to rapid S phase progression impairs the functional and structural characteristics of developing neurons in ASD.
To study the relationship between proliferation and genomic instability in ASD, Wang et al. first compared the replication stress of ASD-derived neural progenitor cells (NPCs) to control patient-derived NPCs. Using a pulse-chase experiment with different thymidine analogs, they discovered that ASD-derived NPCs have more replication forks and origins of replication, which are characteristic of replication stress. Additionally, ASD-derived NPCs have higher levels of global DNA damage markers. To determine the genomic regions most impacted by this replication stress, they performed an assay to identify double-strand breaks upon pharmacologically induced replication stress in NPCs. Wang et al. found that replication stress causes double-strand breaks in a specific subset of neural genes. Since these genes are highly expressed, they then tested for the possibility that the transcription and replication machineries were interfering with one another and causing double-strand breaks. Using a proximity ligation assay, they found high congestion of transcription and replication machinery under replication stress conditions. Analyzing the ontology of these genes, Wang et al. found that they are key structural components for cell-cell adhesion, migration, and dendrite formation. To test the link between these breaks and neurodevelopment, they assayed the ability of NPCs to form rosettes and migrate under replication stress. Rosette formation was disrupted under replication stress and migration from the neurosphere was significantly less. Overall, their work suggests that replication stress induces double-strand breaks in highly expressed genes and that relieving replication stress may be a therapeutic approach early on in ASD.
Proteintech’s HumanKine Noggin (HZ-1118) was used in this study to differentiate Hues6 cells into embryoid bodies for the rosette assay.
About the corresponding author:
Fred Gage, PhD, is the President of the Salk Institute for Biological Studies and the Adler Chair Professor for Research on Age-related Neurodegenerative Disease. His research focus is understanding neurodevelopment.
In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer
Reprogramming of cellular energetics is one of the hallmarks of cancer (PMID 21376230). Visualizing these changes in vivo in patients would advance personalized medicine for diagnosing and treating cancer. In a recent Nature paper, Momcilovic et al. demonstrated that a fluoride radiotracer (18F-BnTP) can be used to monitor the abnormal mitochondrial activity of cancer cells with positron emission tomography (PET) scanning.
To assess its applicability for lung cancers, the authors first examined the biodistribution of 18F-BnTP in a KRAS mouse model. They found that the tracer localizes to lung tumors with a preference for adenocarcinoma (ADC) over squamous cell carcinoma (SCC), suggesting its application as a non-invasive diagnostic. Using pharmacological methods to suppress or enhance oxidative phosphorylation, they showed that tracer uptake is proportional to mitochondrial activity. Combining another tracer for glucose metabolism, 18F-FDG, with 18F-BnTP predicted the efficacy of different drug combinations. Overall, this work suggests that 18F-BnTP may be useful for diagnosing and guiding the treatment of cancer.
Proteintech’s NDUFV1 (11238-1-AP), TIM23 (11123-1-AP), TOM70 (14528-1-AP), and TOM40 (18409-1-AP) antibodies were used in this study in WB to show that the 18F-BnTP signal is a reflection of mitochondrial function and can be used to differentiate ADC’s and SCC’s.
About the corresponding author:
David Shackleford, Ph.D., is an associate adjunct professor in the division of Pulmonary and Critical Care Medicine in the Department of Medicine at the David Geffen School of Medicine at the University of California, Los Angeles. His research focuses on cancer metabolism.
α-Ketoglutarate links p53 to cell fate during tumor suppression
P53 is a famous tumor suppressor and is the most frequently mutated gene across a range of cancers. Although best known for its protective role in the DNA damage response, P53 has also recently been shown to affect cell metabolism. In a recent Nature paper (PMID: 31534224), Morris et al. showed that in pancreatic ductal adenocarcinoma (PDAC), P53 promotes a metabolic profile that inhibits tumor progression.
To determine the role of P53 in cancer metabolism, they modified a PDAC mouse model with a doxycycline-regulated shRNA targeting P53. With doxycycline treatment knocking down P53, tumors progressed rapidly to the malignant state; however, the subsequent restoration of P53 expression in the samples led to decreased tumor size and mortality. Comparing the tumor cells before and after doxycycline removal revealed a change in the metabolic profile characterized by an increase in the alpha-ketoglutarate (aKG) to succinate ratio, a key parameter controlling tumor fate.
In order to demonstrate that increased aKG was the major metabolic mediator of P53 in their model, they added aKG to PDAC cells directly and found it to have the same effects on gene expression and metabolism as restoring P53 expression. This was also observed when increasing aKG levels through the manipulation of Krebs cycle enzymes. Linking this metabolic shift to transcriptional reprogramming, Morris et al. then showed that aKG and P53 levels control the activity of aKG-dependent 5hmc epigenetic writers and that 5hmc levels are negatively correlated with the progression of disease. Thus, this work creates possibilities for novel metabolic approaches for treating PDAC and potentially other cancers that depend on P53 loss for progression.
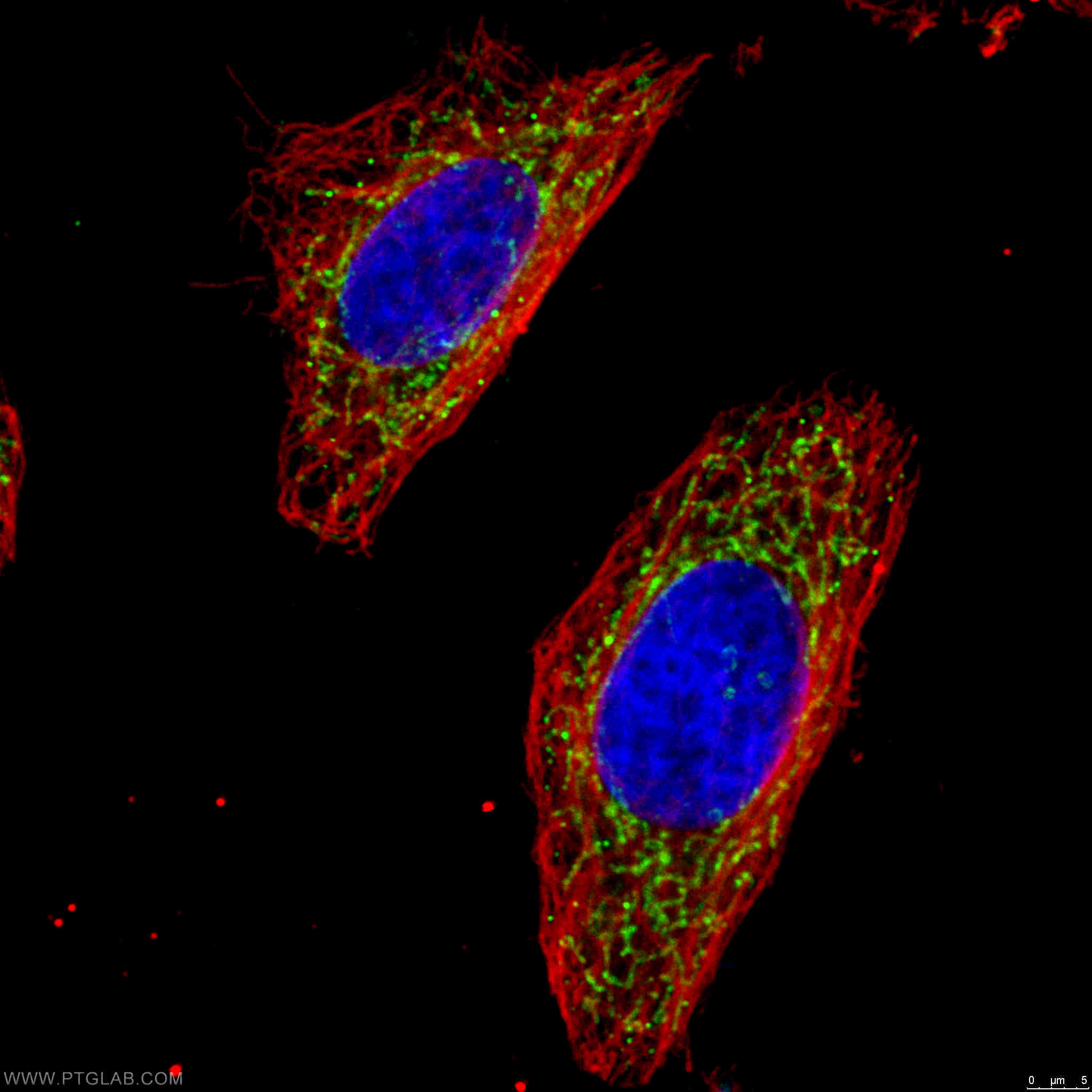
Two Proteintech antibodies were used in this study: IDH1 (12332-1-AP) and OGDH (15212-1-AP). Both antibodies were utilized to confirm the efficiency of the doxycycline-regulated shRNA models used in their major experiments. 12332-1-AP has been validated in WB and ELISA, while 15212-1-AP has been validated in WB, ELISA, and IHC. The specificity of both antibodies has been demonstrated by KD/KO validation.
|
Catalog number: 15212-1-AP KD/KO Validated 18 Publications Immunofluorescent analysis of 4% PFA fixed HeLa cells stained for anti-OGDH (15212-1-AP, 1:50, green) and anti-alpha tubulin (66031-1-IG, 1:100, red). DNA is stained by DAPI (blue). |
 |
About the corresponding author:
Scott Lowe, PhD, is a professor at Memorial Sloan Kettering, chair of the Geoffrey Beene Cancer Research Center, and a Howard Hughes Medical Institute investigator. His work focuses on how the genetics of cancer cells enable tumorigenesis.
12/11/19
Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions.
In a recent paper (PMID: 31523008) Forsström and colleagues examined the molecular causes of mitochondrial myopathies (MM), a class of neuromuscular diseases. The genetic cause of MM is mitochondrial DNA (mtDNA) defects. To better understand the etiology, Forsström et al. focused on the mitochondrial integrated stress response (ISRmt), a major metabolic cellular response to mtDNA mutations and alterations of mtDNA expression. In this study, they analyzed the effects of alterations of transcription factors, regulatory enzymes, and mechanistic target of rapamycin (mTOR) signaling in an in vivo mouse model and in biopsies of MM patients. They discovered that the early stages of the disease were characterized by increased levels of fibroblast growth factor 21 (FGF21) expression, which regulates ISRmt. Knocking out FGF21 resulted in a milder disease phenotype compared to controls in a MM mouse model. Elevated levels of FGF21 was shown to increase glucose uptake, serine de novo biosynthesis, and transsulfuration, which are characteristics of metabolic changes seen in MM, but it had no effect on the accumulation of mtDNA defects. The role of FGF21 depends on cell-type with the greatest impact on muscle cells and the dorsal hippocampus but little in fibroblasts. These findings raise the possibility of targeting FGF21 expression levels and function in individuals affected by MM.
Proteintech’s rabbit anti-MTHFD1L (16113-1-AP) was used in this study to examine the effects of FGF21 on the mitochondrial folate cycle. The authors determined that the expression of MTHFD1L (monofunctional C1-tetrahydrofolate synthase, mitochondrial), an enzyme converting glycine into formate, is regulated by FGF21. MTHFD1L antibody has been validated in Western blot, immunohistochemistry, ELISA, and immunoprecipitation, being specific for human, mouse, and rat.
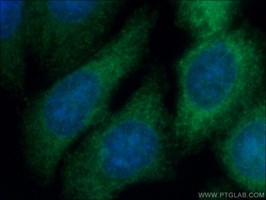
Proteintech’s rabbit anti-CSE (12217-1-AP; Figure 1) was used in this study to examine transsulfuration in MM. The authors have shown that gamma cystathionase (CSE), an enzyme in the transsulfuration pathway, is elevated in MM patients and the MM mouse model and is regulated by FGF21. CSE antibody has been validated in Western blot, immunohistochemistry, ELISA, flow cytometry, immunofluorescence, and immunoprecipitation, being specific for human, mouse, rat, and rabbit.
Proteintech’s rabbit anti-CLPP (15698-1-AP) was used in this study as a marker of mitochondrial unfolded protein response (UPRmt). The authors have shown that caseinolytic mitochondrial matrix peptidase (CLPP), an early marker of UPRmt, is not part of ISRmt. CSE antibody has been validated in Western blot, immunohistochemistry, ELISA, immunofluorescence, and immunoprecipitation, being specific for human, mouse, rat, and zebrafish.
Proteintech's Gamma Cystathionase AntibodyCatalog number: 12217-1-AP KD/KO Validated 88 Publications Figure 1. Immunofluorescent analysis of (-20°C Ethanol) fixed HepG2 cells using 12217-1-AP (Gamma cystathionase antibody) at dilution of 1:50 and Alexa Fluor 488-conjugated AffiniPure Goat Anti-Rabbit IgG(H+L) |
 |
11/27/19
Protein kinase N controls a lysosomal lipid switch to facilitate nutrient signalling via mTORC1.
Mechanistic target of rapamycin (mTOR) signaling, mediated by mTORC1 and mTORC2 protein complexes, regulates a number of vital cellular processes. mTORC1 mainly acts as a nutrient sensor controlling rates of protein synthesis, cell growth, and autophagy.
In a recent Nature Cell Biology paper (PMID: 31451768), Wallroth and colleagues discovered that protein kinase N (PKN2), upon activation via mTORC2, positively regulates mTORC1 nutrient signaling by phosphorylating PI3KC2-β enzyme. PI3KC2-β is responsible for the synthesis of phosphatidylinositol- 3,4-bisphosphate at late endosomes and lysosomes, which then represses mTORC1 activity. However, PI3KC2-β, upon phosphorylation by PKN2, is sequestered in the cytoplasm by binding to 14-3-3 proteins, in turn preventing its inhibitory action on mTORC1 signaling. This signaling pathway was dissected using gain-of-function and loss-of-function cell models (engineered cell lines expressing tagged proteins, transient overexpression of protein mutants, depletion by siRNA, and studying KO cells) coupled with immunoblotting, immunoprecipitation, and immunofluorescence studies. This newly uncovered mechanism of active mTORC2’s influence on mTORC1 signaling represents a parallel route of mTORC1 activation by mitogens, which is classically attributed to AKT activation. Therefore, PKN2 may be an interesting drug target in diseases with altered nutrient signaling, e.g., in cancer or metabolic disorders – diabetes.
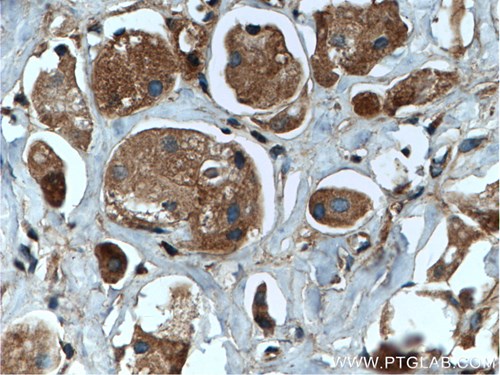
Proteintech’s rabbit anti-Rab35 antibody (11329-2-AP) was used in this study to examine the association of PI3KC2-β with various endosomal Rab GTPases. Rab35 is involved in the rapid recycling of receptors from early endosomes, as well as the transport of receptors from Rab11-positive recycling endosomes. Rab35 antibody has been validated in Western blot, immunohistochemistry, immunofluorescence, and immunoprecipitation, being specific for human, mouse, and rat forms of this protein.
|
Catalog number: 11329-2-AP KD/KO Validated 26 Publications Immunohistochemical analysis of paraffin-embedded human breast cancer tissue slide using 11329-2-AP (RAB35 antibody) at a dilution of 1:200 (under 40x lens) heat mediated antigen retrieved with Tris-EDTA buffer(pH9). |
  |
11/13/19
BCAA catabolism in brown fat controls energy homeostasis through SLC25A44
Warm-blooded, or endothermic, animals use a variety of mechanisms to maintain a constant core temperature. Brown adipose tissue (BAT) is the major organ involved in this process. To generate heat, BAT uncouples oxidative phosphorylation in the mitochondria from ATP generation, resulting in heat rather than stored energy. BAT is thought to exclusively use lipids and carbohydrates as fuel. However, writing in Nature, Yoneshiro et al. suggest that BAT also uses branched-chain amino acids (BCAAs) to generate heat under cold conditions and that inhibiting the use of this fuel source results in diabetes and glucose intolerance.
To gain insight into BAT function, Yoneshiro et al. first determined the metabolic differences between humans with high BAT activity. They found that individuals with high BAT activity have lower BCAA levels after cold exposure than those with low BAT activity. To test whether it is BAT that is responsible for breaking down BCAAs in cold conditions, Yoneshiro et al developed mice that lack the ability to catabolize BCAAs specifically in BAT. Consistent with this hypothesis, they found that BCAA levels do not change in these mice after cold stimulation. Additionally, disabling BCAA metabolism in BAT resulted in glucose intolerance and diet-induced obesity. Using a series of genetic and biochemical experiments, they showed that SLC25A44 is the key transporter of BCAAs into BAT mitochondria. Additionally, mice with knocked-down SLC25A44 have a lower core body temperature and cannot metabolize BCAAs after cold exposure. Overall, Yoneshiro et al.’s work uncovers a critical fuel source of BAT and may lead to insights into obesity and metabolic syndrome.
Proteintech’s anti-TOM20 antibody (11802-1-AP) was used as a mitochondrial marker to show that SLC25A44 is localized to this organelle and as a mitochondrial loading control. This antibody has been tested for WB, IP, IHC, IF, FC, and ELISA, in addition to KD/KO validation.
|
Catalog number : 11802-1-AP KD/KO Validated Citations: 78 Tested Applications: WB, IP, IHC, IF, FC, ELISA Immunofluorescent analysis of ( 4% PFA ) fixed HepG2 cells using 11802-1-AP(TOM20 antibody) at dilution of 1:50 and Alexa Fluor 488-conjugated AffiniPure Goat Anti-Rabbit IgG(H+L) |
 |
10/30/19
The Polycomb Repressor Complex 1 Drives Double-Negative Prostate Cancer Metastasis by Coordinating Stemness and Immune Suppression. (PMID: 31327655)
Prostate cancer is a leading cause of death in men worldwide. Despite the development of powerful immunotherapeutics, the immunosuppressive tumor microenvironment of prostate cancer metastases blunts the therapeutic response. In Cancer Cell, Su and colleagues show that hyperactivity of the polycomb repressor complex (PRC1) increases the expression of chemokines that attract Tregs and promote self-renewal of prostate cancer metastases (PMID 31327655).
Following on from previous work implicating PRC1 in prostate cancer tumorigenesis and metastasis, Su et al. began by comparing PRC1 levels in metastatic prostate cancer to other prostate cancer types and found that PRC1 is upregulated in metastatic samples. To test whether PRC1 is necessary for metastasis, they injected PC3 cells with knocked-down PRC1 into mice and discovered that metastatic lesions were decreased. Mechanistically, they found that PRC1 upregulation in cancer increases CCL2, which promotes stemness and recruits tumor-associated macrophages and Tregs to suppress the immune response. Demonstrating the potential clinical relevance of this result, the inhibition of PRC1 using a small molecule decreased metastasis and improved the efficacy of existing immunotherapeutics. Overall, Su et al.’s work opens the door to novel epigenetic approaches for combating metastasis, while PRC1 hyperactivity may be a common tumorigenic mechanism.
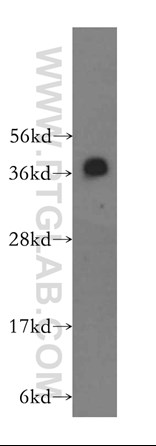
PRC1 is comprised of several subunits, among which RNF2 is critical to its activity. Proteintech’s anti-RNF2 antibody (16031-1-AP) was used to confirm the effectiveness of the investigators’ shRNA constructs targeting this protein and thereby disabling the complex. This antibody has been validated in WB, IP, IHC, IF, and ELISA.
|
Catalog number : 16031-1-AP KD/KO Validated Citations: 3 Tested Applications: WB, IP, IHC, IF, ELISA human brain tissue were subjected to SDS PAGE followed by western blot with 16031-1-AP(RNF2 antibody) at dilution of 1:300 incubated at room temperature for 1.5 hours |
|
Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid (PMID: 31341278)
The clearance of macromolecules from cerebrospinal fluid (CSF) is crucial for the health of the brain. Impaired clearance may play a role in the pathogenesis of Alzheimer’s disease and other neurodegenerative diseases.
The current issue of Nature (PMID: 31341278) highlights the work of Ahn et al. on meningeal lymphatic vessels (mLVs) and their differential involvement in CSF drainage into cervical lymph nodes (cLN). Experimental investigation into mLVs up to this point has been based on dorsal mLVs, which are easily accessible but whose involvement in CSF drainage could not be proven. Gaining access to basal mLVs was more demanding due to their location at the skull base, close to an accumulation of blood vessels and cranial nerves. In this study, Ahn et al. found a method of overcoming these historical experimental limitations to the investigation of basal mLVs and compared the morphology and function of dorsal versus basal mLVs. Histological examinations of dorsal and basal mLVs revealed major differences. Basal mLVs exhibited an abundant number of branches that are similar to functional LVs and far better suited to the uptake of CSF than dorsal mLVs. Basal mLVs display properties of lymphatic pre-collectors due to their lack of SMCs, the mix of button and zipper-like endothelial cell junctions, and the occurrence of valves. In another approach, the authors tracked the lymphatic outflow of the CSF as well as the clearance of macromolecules from the CSF. Starting from the cisterna magna, the signal intensities of both, CSF contrast and macromolecular tracer, peaked in basal mLVs shortly after injection. In dorsal mLVs, the lymphatic outflow peaked relatively late, while uptake of the macromolecular tracer could not be detected, thus emphasizing the importance of basal over dorsal mLVs in CSF clearance.
Many neurodegenerative diseases are related to aging, where reduction of CSF clearance could be connected to mLV remodeling. Deletion of vascular endothelial growth factor 3 (VEGF3, a key player in mLV remodeling) in mice leads to regression of dorsal mLVs before basal mLVs are affected. No impairment of CSF clearance could be detected after regression of dorsal mLVs only; on the contrary, CSF clearance was reduced once VEGF3 deletion started to affect basal mLVs. These observations are an important starting point in tackling the connection between CSF clearance from brain-damaging macromolecules and many age-related pathologies.
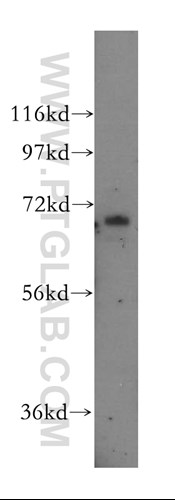
Proteintech’s anti-PROX1 antibody (Cat# 11067-2-AP) was used as an immunofluorescence marker for the lymphatic endothelium in morphological studies comparing dorsal and basal mLVs. This antibody has been validated in Western Blot, immunofluorescence, flow cytometry, ChIP, and ELISA.
|
Proteintech's anti-PROX1 antibody Catalog number : 11067-2-AP KD/KO Validated Citations: 15 Tested Applications: WB, IF, FC, CHIP, ELISA Western blot image: HepG2 cells were subjected to SDS PAGE followed by western blot with 11067-2-AP (PROX1 Antibody) at dilution of 1:600 incubated at room temperature for 1.5 hours. |
 |
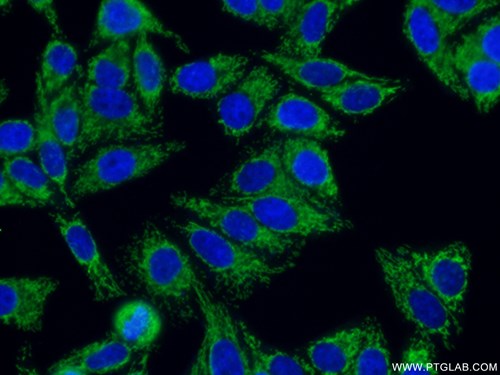
m6A enhances the phase separation potential of mRNA (PMID: 31292544)
Adapting for change is the key job of cells. In the last decade, researchers have discovered a new tool to dynamically influence the transcriptome to fine-tune cellular responses: methylated adenine or N6-methyladenosine (m6a). Advances in sequencing technologies have revealed that this epigenetic mark is abundant throughout the transcriptome and has been implicated in diverse processes including differentiation or the stress-response. The main question in the field is how this small chemical modification can have such powerful effects on the cell.
In a recent Nature paper (PMID: 31292544), Ries et al. show that m6a-RNA can be sequestered by their epigenetic readers into different phase-separated compartments during cell stress.
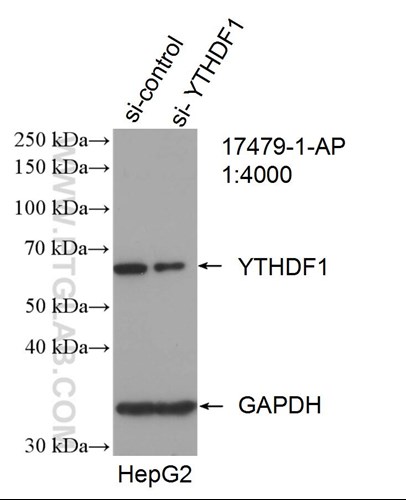
In an elegant set of biochemical experiments, the authors first showed that the m6a-RNA readers, YTHDF1-3, can reversibly separate into a different liquid phase from HEPES buffer by adjusting heat or salt. Afterwards, they found that heat shocks cause this protein to localize to stress granules in a separate liquid phase compartment with the aid of m6a-RNA. Using a METTL14 KO mutant that cannot catalyze m6a markings, they discovered that interaction between m6a and YTHDF1-3 is required for m6a-mRNA transcripts to be concentrated in stress granules during heat and chemical shocks. Ries et al.’s work suggests that m6a may be used to mark transcripts for protection during cellular stress and might be a mechanism for disease pathogenesis and tissue differentiation.
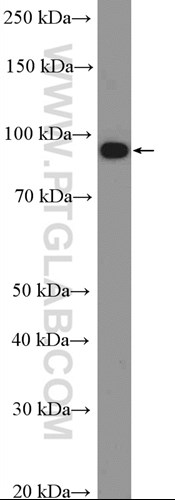
Proteintech’s YTHDF1 (17479-1-AP), YTHDF2 (24744-1-AP), and G3BP1 (13057-2-AP) antibodies were used in this study. 17479-1-AP and 24744-1-AP were used throughout the study to visualize YTHDF1-2 in mouse embryonic stem cells, U2OS, NIH3T3, and HEK293 cells. 13057-1-AP was used as a marker of stress granule formation. All three of these antibodies have been validated by KD/KO experiments to demonstrate specificity.
|
Catalog number: 17479-1-AP KD/KO Validated Citations: 22 Tested Applications: WB, IP, IF, IHC, ELISA Western blot image: |
 |
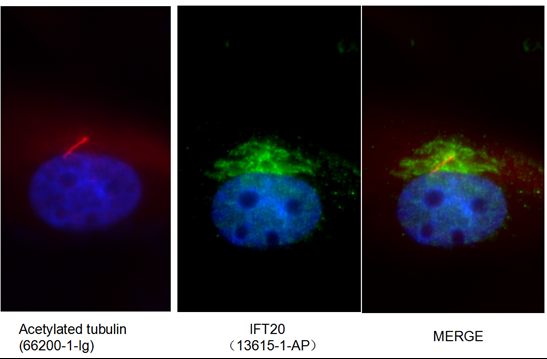
Hypomorphic mutations of TRIP11 cause odontochondrodysplasia. (PMID: 30728324)
The molecular causes of many human genetic diseases are yet to be identified. In a recent paper (PMID: 30728324), Wehrle and colleagues show that odontochondrodysplasia (ODCD), a form of skeletal dysplasia, is caused by hypomorphic mutations in the TRIP11 gene. TRIP11 encodes GMAP-210 (Golgi-associated microtubule-binding protein 210) which regulates protein transport and posttranslational modification within the Golgi apparatus. Severely reduced levels of GMAP-210 in patients’ cells disrupt the structure of the Golgi apparatus and abnormal glycosylation of cargo proteins, which negatively impacts chondrocyte differentiation.
The authors investigated posttranslational modifications of protein using western blotting and analyzing band patterns of glycosylated protein species and the resulting shifts of their molecular weights. Immunofluorescence imaging was used to look at the co-localization studies of Golgi proteins and IFT20, a binding partner of GMAP-210 that localizes both to the Golgi and primary cilium.
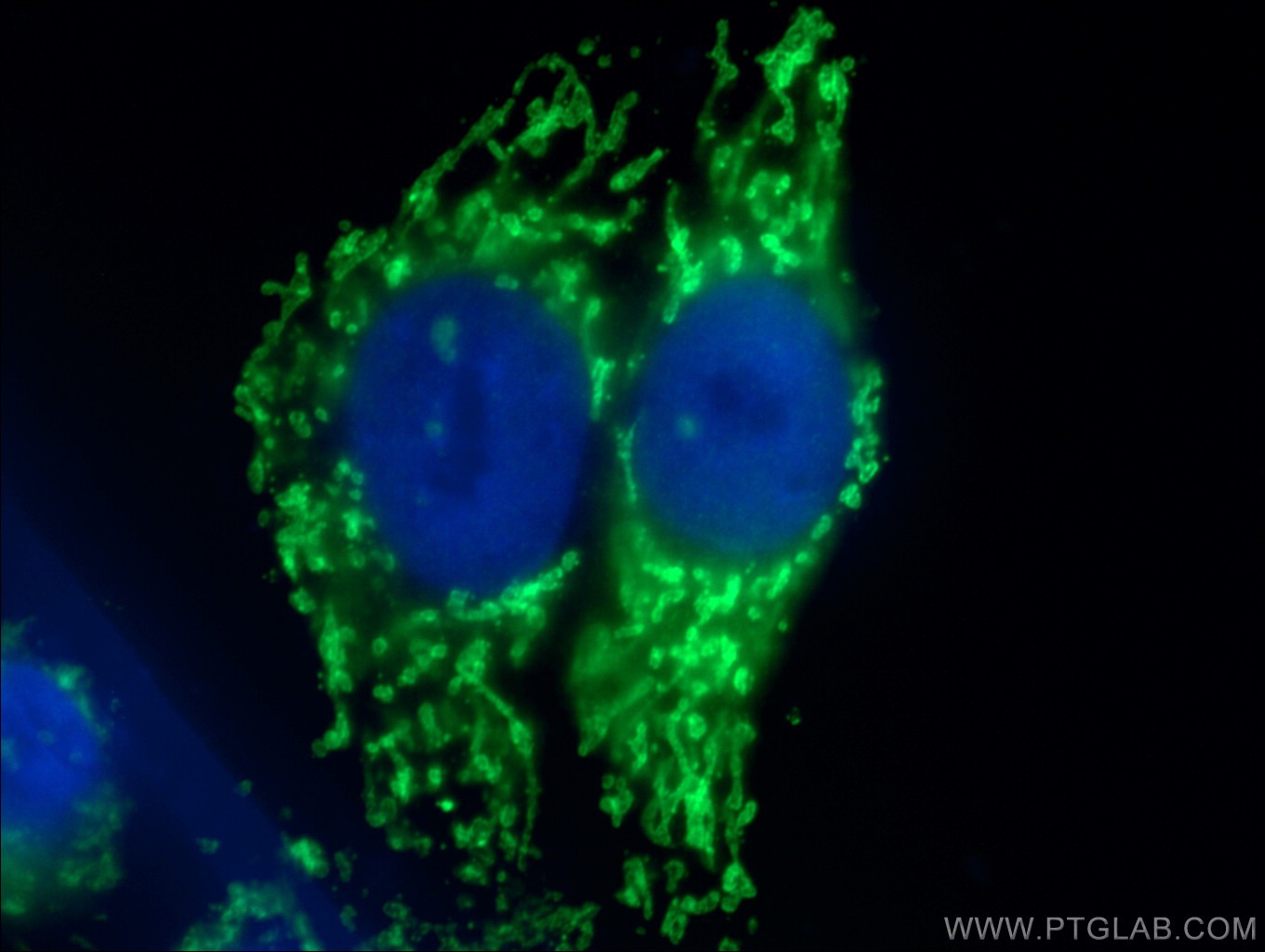
Proteintech’s rabbit anti-IFT20 antibody (13615-1-AP) was used in this study to show that GMAP-210 protein mediates correct subcellular localization of IFT20 to the Golgi apparatus. This antibody has been validated in western blot, immunohistochemistry, and chromatin immunoprecipitation with genetic knockdown data to demonstrate its specificity.
|
KD/KO Validated Catalog number: 13615-1-AP Citations: 31 Tested Applications: WB, IP, IHC, IF, ELISA IF image: Immunofluorescent analysis of (10% Formaldehyde) fixed MDCK cells using 13615-1-AP (IFT20 antibody) at dilution of 1:100 and Alexa Fluor 488-conjugated AffiniPure Goat Anti-Rabbit IgG(H+L) |
|
|
Reviews: 5 star Verified customer: "Excellent for IF labelling of the Golgi under PFA fixation." |
For more cilia-related antibodies, please see our cilia card.
Additionally, Proteintech’s rabbit anti-decorin antibody (14667-1-AP) was used to look at posttranslational modifications of extracellular matrix components.
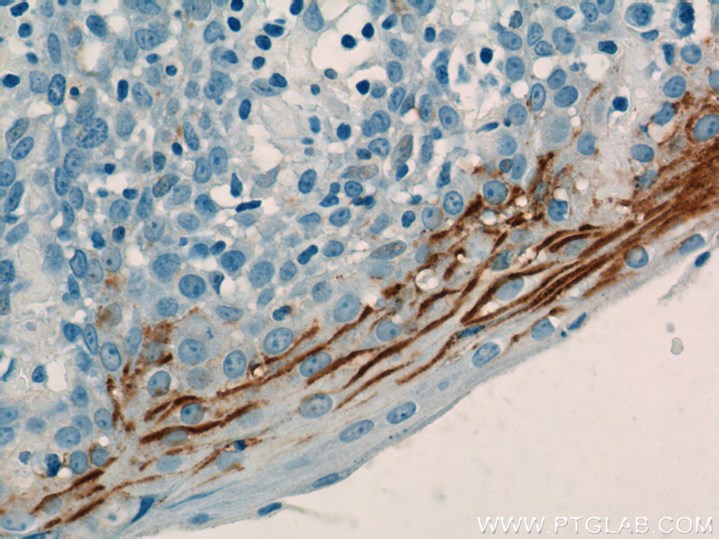
Single-Cell Analysis of the Liver Epithelium Reveals Dynamic Heterogeneity and an Essential Role for YAP in Homeostasis and Regeneration.
The liver plays a crucial role in protein, carbohydrate, and lipid metabolism, the detoxification of drugs, and the breakdown of hormones. Two cell types, hepatocytes and biliary epithelial cells (BECs), are critical to the regenerating capabilities of the liver.
The current issue of Cell Stem Cell (Vol. 25, issue 1) features the work of Pepe-Mooney and coworkers who elucidated the dual role of the YAP signaling pathway in liver cell homeostasis and the initiation of regenerative processes (PMID: 31080134). YAP is a transcription factor that is involved in the regulation of cell proliferation as well as apoptosis. In an elegant assay, the authors utilized FACS followed by a single-cell RNA-seq to compare the expression profiles of 2,344 homeostatic BECs. Upon the differential expression of several YAP target genes, they identified two major cell clusters, YAP activated and non-activated cells. To identify the reason for this heterogeneity in YAP expression, the assay was repeated for BECs and hepatocytes after drug-induced mimicking of chronic liver damage in vivo. The results revealed the importance of YAP expression in the regenerative response of BECs and hepatocytes under liver-damaging conditions. The exposure of BECs to physiological levels of bile acid (BA) revealed the additional role of the YAP signaling pathway in cell homeostasis, dependent on intracellular BA uptake. This study presents a great basis for further investigations to elucidate cell type-dependent roles and the complexity of YAP signaling.
Beside the two major clusters, the initial RNA-seq screening revealed a small subset of cells co-expressing Dmbt1 and Ly6d, which are markers for extrahepatic BECs.
Proteintech’s Rabbit anti-LY6D (Cat# 17361-1-AP) was used for immunofluorescence staining as a differential marker for extrahepatic BECs. Based on the antibody performance it was possible to separate extrahepatic from intrahepatic BECs. This antibody has additionally been validated in Western Blot, immunohistochemistry, and ELISA.
|
Catalog number: 17361-1-AP Citations: 1 Tested Applications: WB, IHC, ELISA IHC image: Immunohistochemical analysis of paraffin-embedded human tonsillitis tissue slide using 17361-1-AP( LY6D Antibody) at dilution of 1:50 (under 40x lens) |
 |
Maintenance of CTCF- and Transcription Factor-Mediated Interactions from the Gametes to the Early Mouse Embryo
Is a child being born to a home with food insecurity? A frigid climate? A place with high toxic exposure? Being able to endow children with genetic adaptations to the environment provides significant evolutionary fitness. The prevailing notion is that these tweaks in the gamete stage are mediated by methylation and histone modifications. In the cover story for July’s Molecular Cell (PMID: 31056445), Jung et al. suggest that transcription factor sites and enhancer loops are also inherited from the parents.
Using a transposon accessibility assay and ChIP-seq, they show that mature sperm have transcription machinery and enhancer loops at many sites, but that the corresponding genes are not actively transcribed until embryogenesis. Comparing the loops of DNA in the parental gametes and embryos, they further showed that many of the CCCTC-binding factor (CTCF) and other transcription factor loops are inherited in a parent-specific manner. Their work introduces chromatin structure and early transcription factor priming as novel parental imprinting mechanisms for further investigation.
Proteintech’s Znf143 antibody (16618-1-AP) was used in this study to show that Zinc-finger transcription factors are present at many of inherited sites. This antibody has been validated in Western blot, immunohistochemistry and chromatin immunoprecipitation with genetic knockdown data to demonstrate its specificity.

Feifan Guo. Professor Guo is a professor at the Shanghai Institute of Nutrition and Health, and her work focuses on the molecular mechanisms underlying nutrition-related metabolic diseases.
- 32727915