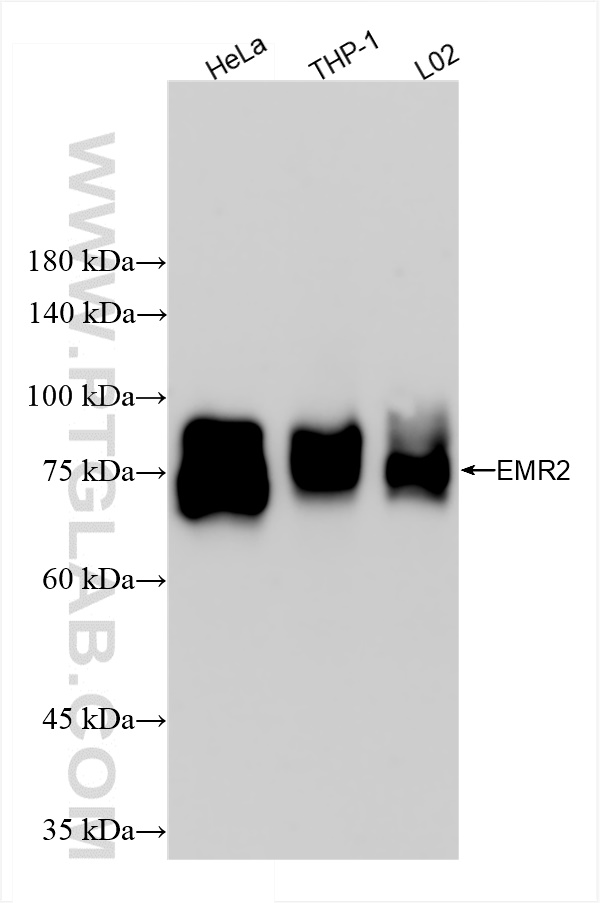
验证数据展示
经过测试的应用
| Positive WB detected in | HeLa cells, THP-1 cells, L02 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:2000-1:10000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
84608-5-RR targets EMR2 in WB, ELISA applications and shows reactivity with human samples.
| 经测试应用 | WB, ELISA Application Description |
| 经测试反应性 | human |
| 免疫原 | Recombinant protein 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Recombinant |
| 产品类型 | Antibody |
| 全称 | egf-like module containing, mucin-like, hormone receptor-like 2 |
| 别名 | EGF-like module receptor 2, EGF like module receptor 2, CD312, Adhesion G protein-coupled receptor E2, ADGRE2 |
| 计算分子量 | 90 kDa |
| 观测分子量 | 75~90 kDa |
| GenBank蛋白编号 | NM_013447.3 |
| 基因名称 | EMR2 |
| Gene ID (NCBI) | 30817 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Protein A purfication |
| UNIPROT ID | Q9UHX3-1 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
EMR2 (Epidermal Growth Factor-like module-containing Mucin-like Hormone Receptor 2), also known as ADGRE2, is a member of the adhesion-GPCR family that contains extracellular EGF-like domains. EMR2 contains a total of five tandem EGF-like domains and expresses similar protein isoforms consisting of various numbers of EGF-like domains as a result of alternative RNA splicing (PMID: 10903844). EMR2 is restricted to myeloid cells including monocytes, macrophages, dendritic cells and granulocytes28 and its expression is highly regulated during monocyte/macrophage differentiation (PMID: 31969668).The ligation of EMR2 could increase neutrophil adhesion, migration and anti-microbial mediator production and could enhance the systemic inflammation (PMID: 27905560).
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for EMR2 antibody 84608-5-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |