验证数据展示
经过测试的应用
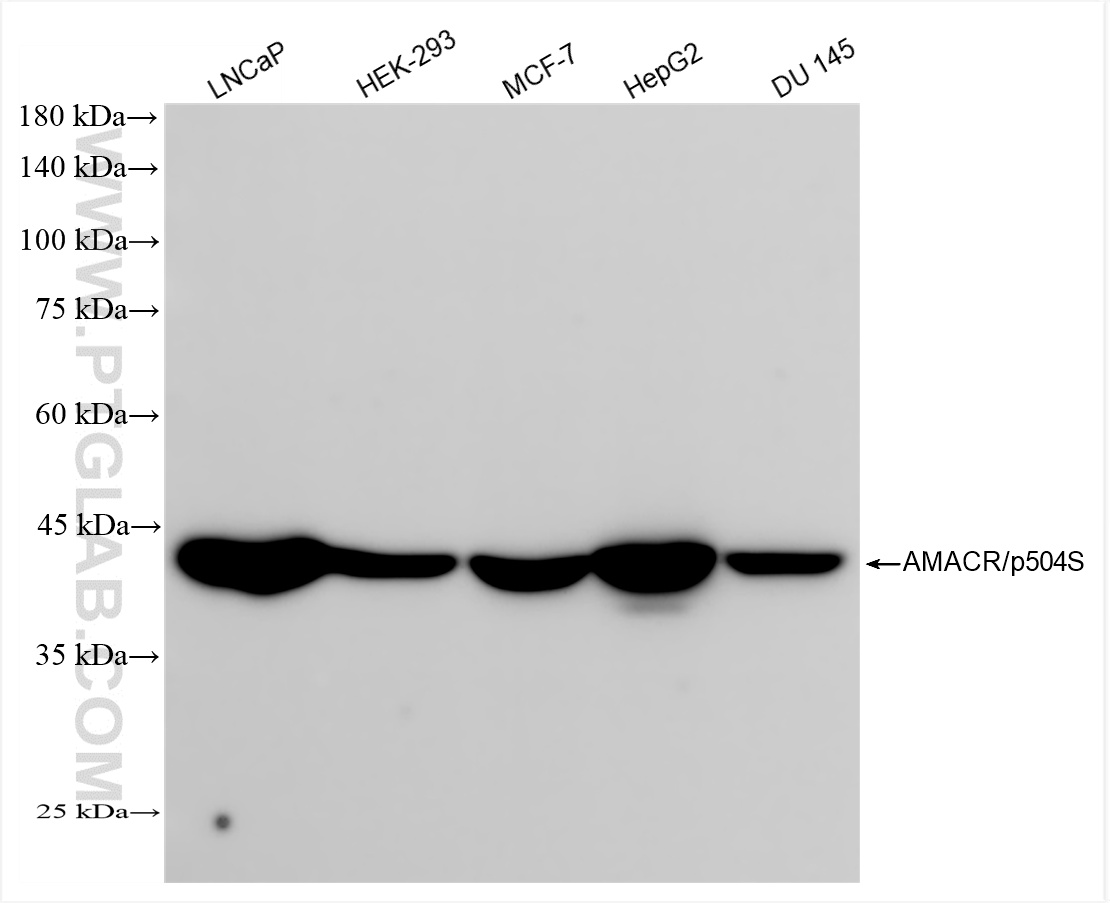
| Positive WB detected in | LNCaP cells, HEK-293 cells, MCF-7 cells, HepG2 cells, DU 145 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
84891-2-RR targets AMACR/p504S in WB, ELISA applications and shows reactivity with human samples.
| 经测试应用 | WB, ELISA Application Description |
| 经测试反应性 | human |
| 免疫原 | AMACR/p504S fusion protein Ag8720 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Recombinant |
| 产品类型 | Antibody |
| 全称 | alpha-methylacyl-CoA racemase |
| 别名 | AMACR, p504S, EC:5.1.99.4, Alpha-methylacyl-CoA racemase, 2-methylacyl-CoA racemase |
| 计算分子量 | 382 aa, 42 kDa |
| 观测分子量 | 42 kDa |
| GenBank蛋白编号 | BC009471 |
| 基因名称 | AMACR |
| Gene ID (NCBI) | 23600 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Protein A purification |
| UNIPROT ID | Q9UHK6 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
AMACR(Alpha-methyl acyl-CoA racemase) belongs to the CaiB/BaiF CoA-transferase family. A mitochondrial and peroxisomal enzyme catalyzes the conversion of 2R stereoisomers of phytanic and pristanic acid to their S counterparts. AMACR has previously been shown to be a highly sensitive marker for colorectal and clinically localized prostate cancer (PCa). However, AMACR expression is down-regulated at the transcript and protein level in hormone-refractory metastatic PCa, suggesting a hormone-dependent expression of AMACR(PMID:12213712). It has 3 isoforms produced by alternative splicing and can form a dimer of 70-75 kDa(PMID:21812041). Defects in AMACR cause alpha-methyl acyl-CoA racemase deficiency (AMACRD) and congenital bile acid synthesis defect type 4 (CBAS4).
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for AMACR/p504S antibody 84891-2-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |