ATAC-Seq Kit
ATAC-Seq-Kit-AM1301
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
ATAC-Seq Kit Overview
Looking for bulk recombinant Tn5 transposase? Get a quote.
Need to multiplex more than 16 samples? See our Nextera™-Compatible Multiplex Primers (96 plex).
ATAC-Seq is a rapid assay that allows analysis of epigenetic profiles across the genome by identification of regions that have open or accessible chromatin states. Because of the assay’s speed, simplicity, and applicability to a wide range of sample types, ATAC-Seq has become a commonly-used epigenetic assay, and it can serve as a gateway to further, more detailed epigenetic analyses.
In the ATAC-Seq assay, intact nuclei from cell or tissue samples are treated with a hyperactive Tn5 transposase mutant. This transposase is able to simultaneously tag the target DNA with sequencing adapters and fragment the DNA in a process termed tagmentation.
The ATAC-Seq Kit from Active Motif provides the reagents necessary to produce 16 unique sequencing-ready Illumina®-compatible ATAC-Seq libraries from 20 – 30 mg tissue or 50,000 – 100,000 cells per reaction. The optimized protocol guides you through the steps for sample preparation, tagmentation and library preparation, yielding next-gen sequencing-ready libraries that can be multiplexed in a single flow cell sequencing run.
ATAC-Seq Kit Highlights:
- Reveal the genomic sequence of open chromatin regions
- Yields next-gen sequencing-ready libraries in hours
- Simple and rapid optimized protocol & reagents
Contents
ATAC-Seq Kit Contents
The ATAC-Seq Kit is shipped at two temperatures, with one box on dry ice for components to be stored at -20°C, and a second box at room temperature for components to be stored at 4°C. Please store components according to the storage conditions in the manual after first use. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
Reagents included:
- ATAC Lysis Buffer, store at RT
- Assembled Transposomes, store at -20°C
- 2X Tagmentation Buffer, store at -20°C
- 1X PBS, store at -20°C or RT
- 10X PBS, store at RT
- 10% Tween 20, store at RT
- 1% Digitonin, store at -20°C
- DNA Purification Columns, store at RT
- DNA Purification Binding Buffer, store at RT
- DNA Purification Wash Buffer, store at RT
- DNA Purification Elution Buffer, store at RT
- 3 M Sodium Acetate, store at RT
- 10 mM dNTPs, store at -20°C
- 5X Q5 Buffer, store at -20°C
- Q5 High-Fidelity DNA Polymerase, store at -20°C
- i7 Indexed Primer 1, store at -20°C
- i7 Indexed Primer 2, store at -20°C
- i7 Indexed Primer 3, store at -20°C
- i7 Indexed Primer 4, store at -20°C
- i5 Indexed Primer 1, store at -20°C
- i5 Indexed Primer 2, store at -20°C
- i5 Indexed Primer 3, store at -20°C
- i5 Indexed Primer 4, store at -20°C
- SPRI Beads, store at 4°C
Data
ATAC-Seq Kit Data
The ATAC-Seq kit contains the necessary reagents and enzymes to perform 16 ATAC-Seq reactions. These kit components are optimized to work efficiently and have been validated to generate high-quality ATAC-Seq data.
Reveal Open Chromatin Locations at Promoters
The chromatin at promoters of genes that are transcriptionally active are generally open or accessible. ATAC-Seq assays result in peaks at these locations, revealing those open chromatin regions in the samples being investigated.
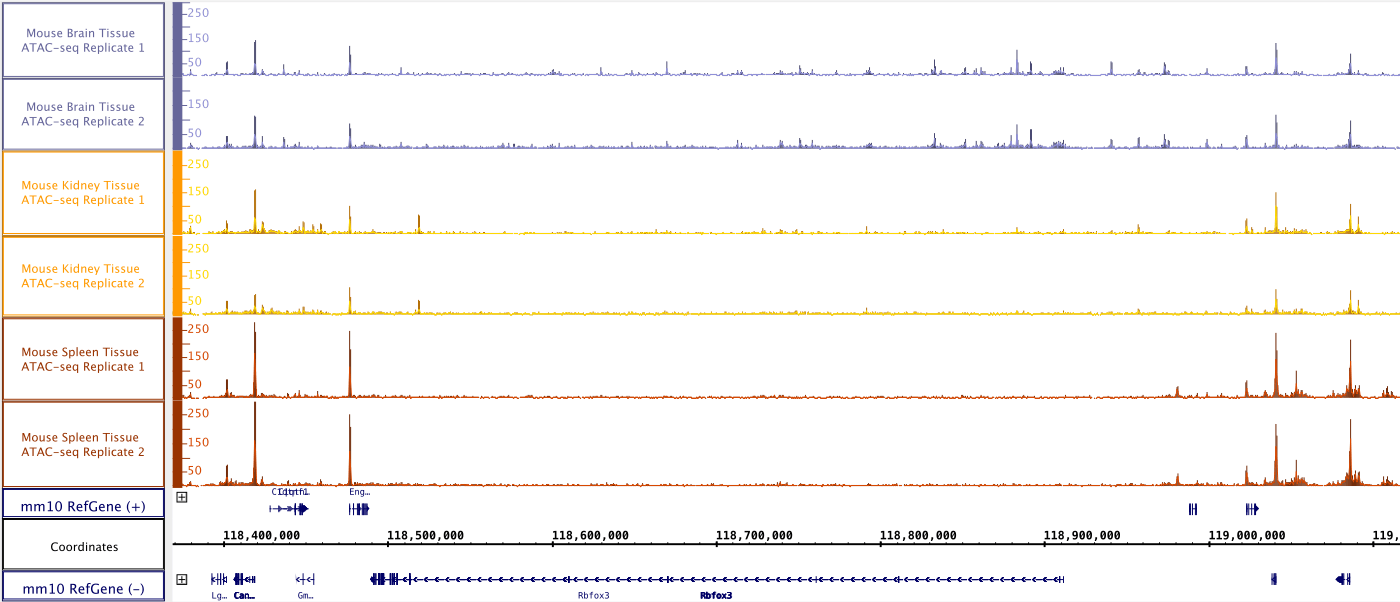
Figure 1: Genome Browser Display of ATAC-Seq Kit results. ATAC-Seq Kit was used to assess the genome-wide open chromatin state of mouse brain tissue samples. The genome browser tracks shown here are at the position of the FOXP3 gene, which encodes a marker of post-mitotic neurons. The peaks indicate open chromatin signal at the promoter of the FOXP3 gene.
Kit Performance Correlates with Published Data
The Active Motif ATAC-Seq Kit generates high-quality data. ATAC-Seq data from experiments performed with the ATAC-Seq Kit correlates very well with published ATAC-Seq data, highlighting that this assay is robust and reproducible.
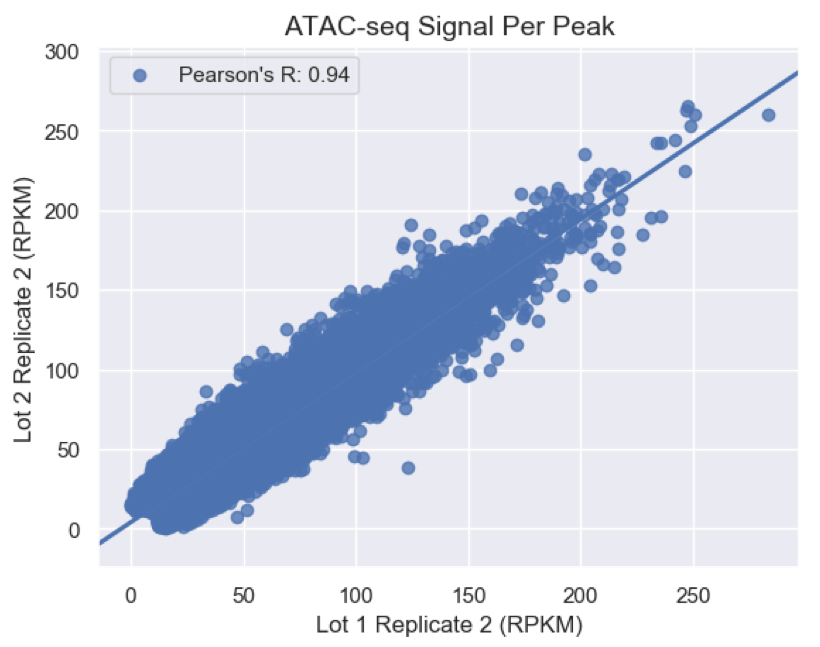
Figure 2: ATAC-Seq Kit Libraries Correlation with Published ATAC-Seq Dataset. Here, the correlation between RPKM signal in called peaks of published ATAC-Seq data (Buenrostro, J. D. et al. (2013) Nat. Methods 10: 1213-1218) and a dataset generated with the Kit in GM12878 cells is shown, indicating a tight correlation with a score of 0.94.
What our customers are saying about our ATAC-Seq kit:
FAQs
ATAC-Seq Kit FAQs
Is this kit the same as the original Buenrostro et al. Nature 2013 paper?
This kit is based off a combination of the original Buenrostro paper and the Corces et al. Nature 2017 paper, and we have optimized the buffers for a more robust and efficient assay.
What is the source of the recombinant Tn5 enzyme in this kit?
The recombinant Tn5 enzyme in this kit is a hyperactive mutant that we cloned, expressed, purified, and test in-house. The Assembled Transposomes containing this enzyme are provided pre-loaded with the next-gen sequencing adapters, so it is ready-to-use without any optimization or other preparation required.
What is the minimum number of cells or tissue per reaction?
50,000 cells or 20 mg tissue.
Can frozen tissue and cells be used?
Yes, but the samples must be high quality to preserve viability. Cells should be cryopreserved in a controlled-rate freeze with media formulated to protect against ice crystal formation and subsequent cell damage, and tissue should be flash frozen at -80°C.
Can nuclei be frozen for later use in ATAC-Seq?
Yes, extracted nuclei can be cryopreserved and frozen at -80°C for later use. Nuclei can also be pooled together to increase the sample size.
Can FACS-sorted cells be used?
It is possible, but cells could be damaged during the FACS-sorting process. For good ATAC-Seq libraries, cells must be viable.
Do you have a recommendation for a 40 micron mesh strainer for tissue samples?
Yes, we recommend the Falcon 40 micron cell strainer (Falcon catalog no. 352340).
Is DNase treatment recommended for all samples?
DNase treatment can help in certain situations such as mitochondrial DNA depletion, or if there are 15% dead cells in the sample, but it can end up removing cells you want data from if the cells aren’t healthy and able to exclude the enzyme. It’s only recommended as an option when cells are viable and won’t also be digested by the enzyme.
I don’t have a thermomixer. Can I still use the kit?
Using a thermomixer is recommended. However, we have tested this and incubated the tagmentation reaction without the use of a mixer at 37°C in a thermal cycler with a heated lid and the reaction was successful.
Can all the reagents and kit components, including master mixes and enzymes, thaw out and be prepared at room temperature?
Everything except the Assembled Transposomes and Q5 DNA polymerase can be thawed at room temperature. The Assembled Transposomes and Q5 DNA Polymerase are formulated with glycerol and will not need thawing, just put these on ice. The other components can be frozen again and thawed again for future reactions, so not all 16 reactions need to be done simultaneously.
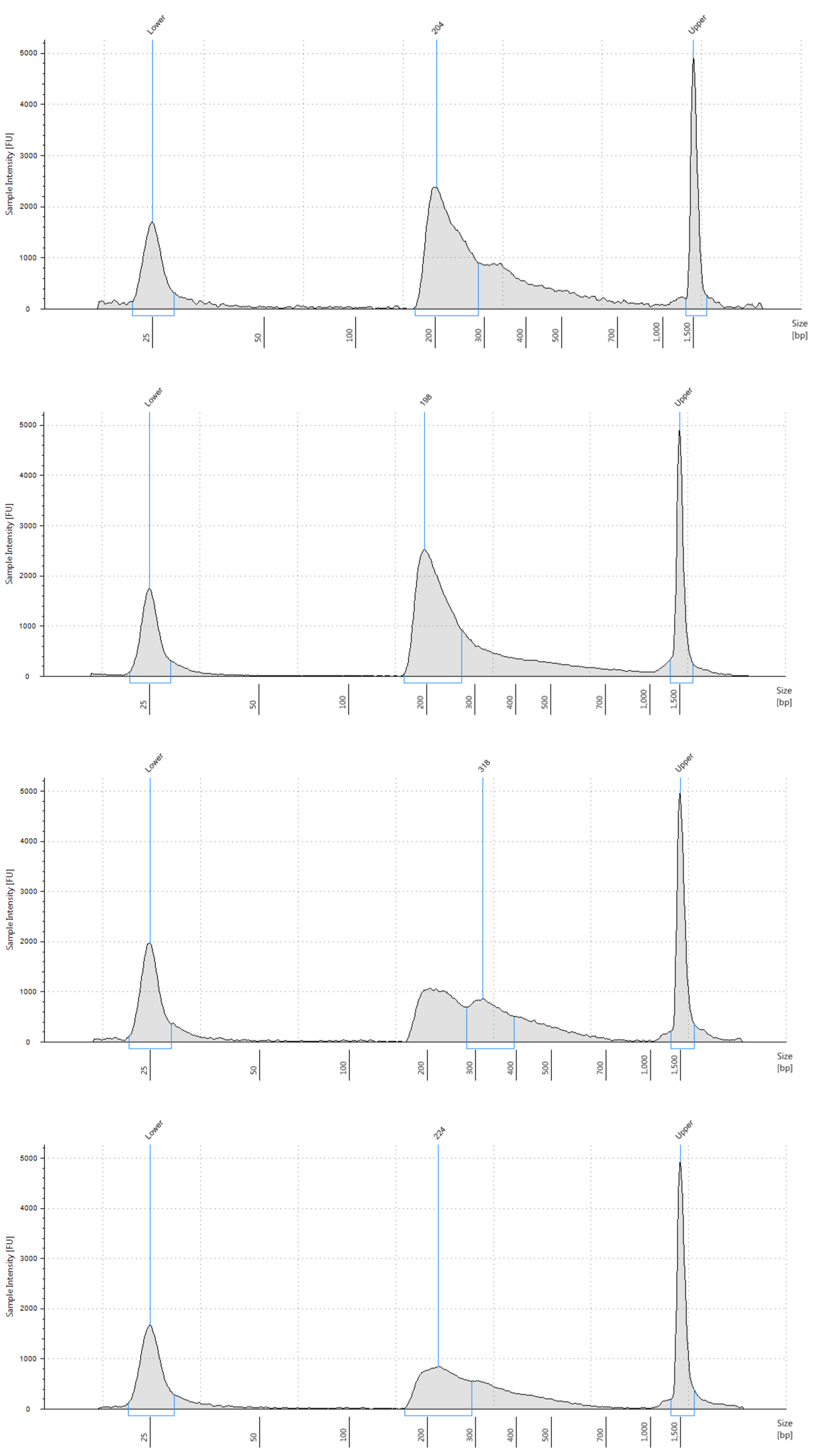
What is the expected final library size, and what should the library traces look like on a fragment analyzer?
How many sequencing reads are needed per sample?
Typically 30 million paired-end reads is sufficient, with 20 million being a minimum. The number of reads is dependent both on the sample genome and the end goal of the ATAC-Seq assay. If the sample genome is very large, or if more advanced analysis is required, more sequencing reads may be needed.
Can I multiplex more than 16 samples using the ATAC-Seq Kit?
The ATAC-Seq Kit is supplied with 4x4 unique dual indexes for 16 unique samples. The indexed primers in the kit are identical to the Illumina Nextera primers corresponding to N701-N704 and N501-N504. If you would like to multiplex more than 16 samples our Nextera™-Compatible Multiplex Primers (96 plex) kit (Cat. No. 53155) enables multiplexing up to 96 reactions. These primers are provided at a concentration of 25 µM to be used directly in our Kits. You could also purchase and combine other Illumina Nextera primers at the same concentration (25 µM) as those in the kit.
Questions about sample library QC? Read more on our blog.
Documents
ATAC-Seq Kit Documents