ChIP-IT High Sensitivity®
ChIP-IT-High-Sensitivity-AM868
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
The ChIP-IT High Sensitivity® (HS) Kit is the most sensitive ChIP kit available on the market. It is designed to allow high quality DNA enrichment from as little as 1,000 cells per ChIP reaction for abundant targets. The kit is also optimized to give superior results when performing chromatin immunoprecipitation using antibodies against low abundance proteins, such as transcription factors, or antibodies with sub-optimal binding affinities. To see how these features compare to the other ChIP Kits we offer, see our ChIP Kit Selection Guide.
Higher signal and lower non-specific DNA pulldown is achieved through the incorporation of specialized protein G agarose beads and optimized fixation, binding, and wash buffers. In addition, washes are performed via gravity filtration, which is faster, easier, and more consistent than magnetic bead capture. The ChIP-IT High Sensitivity® (HS) Kit has also been validated for use in ChIP-Seq, even when using antibodies that target low abundance transcription factors.
ChIP-IT High Sensitivity® Advantages
- Ideal for low abundance transcription factors or antibodies with low binding affinities
- Sensitive enrichment of DNA from as little as 1,000 cells per immunoprecipitation reaction for high abundance target proteins and as little as 50,000 cells for low abundance transcription factors
- Optimized reagents reduce background levels caused by non-specific binding events
- Filtration based washes offer a faster, easier solution with better consistency than magnetic capture
- Highly robust procedure has been validated across multiple sample types with proven performance in both qPCR and ChIP-Seq analysis
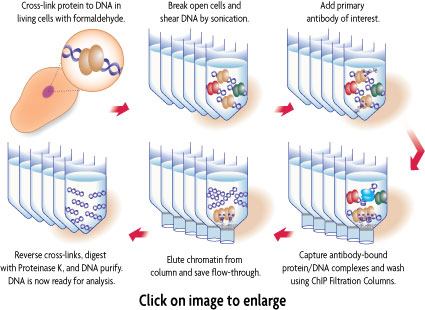
Figure 1: Schematic of chromatin immunoprecipitation using ChIP-IT High Sensitivity.
In ChIP-IT High Sensitivity®, intact cells are fixed with a specially formulated formaldehyde buffer, which cross-links and preserves protein/DNA interactions. DNA is then sheared into small fragments using sonication and incubated with an antibody directed against the DNA-binding protein of interest. The antibody-bound protein/DNA complexes are immunoprecipitated through the use of Protein G agarose beads and washed via gravity filtration. Following immunoprecipitation, the DNA cross-links are reversed, the proteins are removed by Proteinase K and the DNA is recovered and purified. ChIP enriched DNA can be used for either gene-specific or whole-genome analysis.
Active Motif also offers products compatible with the ChIP-IT High Sensitivity Kit:
- ChIP-IT® qPCR Analysis Kit – simplifies qPCR data analysis and enables normalization across multiple sample types and experiments
- High Sensitivity Chromatin Preparation – designed to isolate formaldehyde fixed chromatin from cultured cells or tissue samples for use in ChIP
- qPCR Primer Sets for a variety of popular gene targets in a number of model organisms.
- Antibodies validated with our ChIP-IT High Sensitivity Kit.
In addition, our Epigenetic Services can perform ChIP, ChIP-Seq and many other genome-wide data generation and analysis services for you.
Contents
ChIP-IT High Sensitivity® Contents & Storage
Please note that the ChIP-IT High Sensitivity Kit and High Sensitivity Chromatin Kit are shipped on dry ice and contains reagents with multiple storage temperatures inside. Please store each component at the temperature indicated below. All reagents are guaranteed stable for 6 months from date of receipt when stored properly. Do not re-freeze the Protein G Agarose Beads after you have received them.
The High Sensitivity Chromatin Kit is a companion module to our ChIP-IT High Sensitivity Kit and contains just those reagents required for optimizing the fixation, shearing, and lysis conditions before proceeding to the full scale ChIP reaction.
ChIP-IT High Sensitivity Kit
- RNase A (10 µg/µl); Store at -20°C
- Proteinase K (10 µg/µl); Store at -20°C
- 100 mM PMSF; Store at -20°C
- Protease Inhibitor Cocktail (PIC); Store at -20°C
- Precipitation Buffer; Store at -20°C
- Carrier; Store at -20°C
- 10X PBS; Store at -20°C
- Blocker; Store at -20°C
- Fixation Buffer; Store at 4°C
- Protein G Agarose beads; Store at 4°C
- Stop Solution; Store at RT
- Chromatin Prep Buffer; Store at RT
- ChIP Buffer; Store at RT
- Wash Buffer AM1; Store at RT
- Elution Buffer AM4; Store at RT
- ChIP Filtration Columns; Store at RT
- 5 M NaCl; Store at RT
- TE, pH 8.0; Store at RT
- Detergent; Store at RT
- DNA Purification Binding Buffer; Store at RT
- 3 M Sodium Acetate; Store at RT
- DNA Purification Wash Buffer; Store at RT
- DNA Purification Elution Buffer; Store at RT
- DNA Purification Columns; Store at RT
High Sensitivity Chromatin Prep Kit
- RNase A (10 µg/µl); Store at -20°C
- Proteinase K (10 µg/µl); Store at -20°C
- 100 mM PMSF; Store at -20°C
- Protease Inhibitor Cocktail (PIC); Store at -20°C
- Precipitation Buffer; Store at -20°C
- Carrier; Store at -20°C
- 10X PBS; Store at -20°C
- Fixation Buffer; Store at 4°C
- Swelling Buffer; Store at 4°C
- Stop Solution; Store at RT
- Chromatin Prep Buffer; Store at RT
- ChIP Buffer; Store at RT
- DNA Purification Elution Buffer; Store at RT
- 5 M NaCl; Store at RT
- TE, pH 8.0; Store at RT
- Detergent; Store at RT
Data
ChIP-IT High Sensitivity® Data
Detect Low Abundance Protein Targets
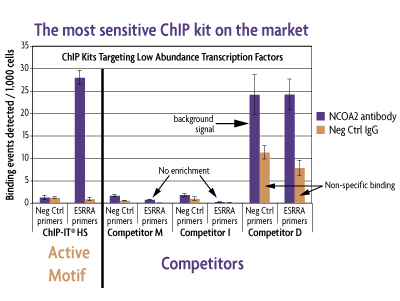
The ChIP-IT High Sensitivity® Kit is ideal for use with challenging antibodies that do not give signal with other ChIP methods, as the method is sensitive enough to detect specific binding of even low abundance proteins. Below is a comparison of ChIP-IT High Sensitivity with other commercially available ChIP kits. ChIP was performed using untreated MCF-7 chromatin and an antibody that binds the low abundance nuclear receptor co-activator 2 (NCOA2) transcriptional co-factor. It was also performed with a negative control IgG and a negative control primer set (Active Motif Catalog No. 71001) to monitor non-specific binding. Only the ChIP-IT High Sensitivity method was able to detect NCOA2 binding at specific genomic locations (ESRRA promoter), providing approximately 20-fold higher enrichment than the negative controls. Although ESRRA shows qPCR signals using the kit from Competitor D, the enrichment levels are poor due to the high background from the negative control primer set and the non-specific binding detected with the IgG.
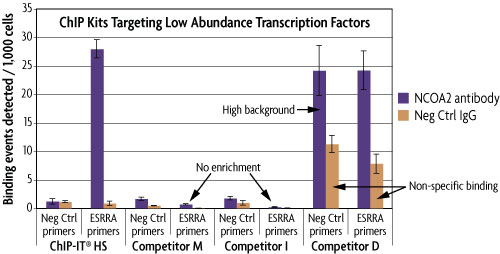
Figure 2: Comparison of ChIP kits targeting a low abundance transcription factor.
MCF-7 chromatin was prepared according to each manufacturer's recommendations for their ChIP assay from 1.5 x 106 cells. ChIP was performed with the optimal amount of chromatin as suggested by the protocol for each manufacturer using an antibody for the low abundance NCOA2 protein and a negative control IgG. Following enrichment, qPCR was performed using the ChIP-IT qPCR Analysis Kit (Catalog No. 53029) in order to normalize the data and allow for direct comparison of the results. Data is expressed as binding events detected per 1,000 cells which represents the average of the raw data triplicates adjusted for the amount of chromatin in the reaction, the resuspension volume and the primer efficiency.
Achieve Greater Sensitivity
A comparison of ChIP-IT High Sensitivity to other commercially available ChIP Kits when using the high-quality H3K4me3 antibody (Catalog No. 39915) shows specific signal for the positive control GAPDH promoter (Catalog No. 71006) using ChIP-IT HS, Competitor M, Competitor I, and Competitor D's Kits. Due to the optimized conditions with the ChIP-IT High Sensitivity, the detectable enrichment was 2-3 fold higher than the enrichment obtained from Competitor M and Competitor I. Only Competitor D did not show good enrichments as a result of high non-specific binding as seen with the signal from the negative control qPCR primer set and the high signal in the IgG reactions.
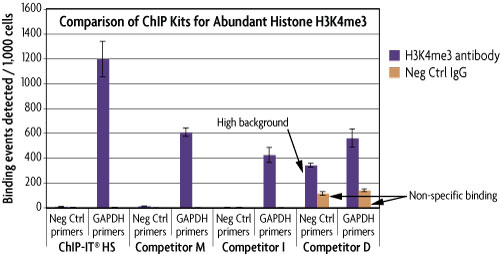
Figure 3: ChIP-IT High Sensitivity shows better enrichment than competitor ChIP Kits.
MCF-7 chromatin was prepared according to each manufacturer's recommendations for their ChIP assay from 1.5 x 106 cells. ChIP was performed with the optimal amount of chromatin as suggested by the protocol for each manufacturer using an antibody for the abundant Histone H3K4me3 (Catalog No. 39915) and a negative control IgG. Following enrichment, qPCR was performed using the ChIP-IT qPCR Analysis Kit (Catalog No. 53029) in order to normalize the data and allow for direct comparison of the results. Data is expressed as binding events detected per 1,000 cells which represents the average of the raw data triplicates adjusted for the amount of chromatin in the reaction, the resuspension volume and the primer efficiency.
Use Less Sample Material
The optimized ChIP buffers included in the ChIP-IT High Sensitivity Kit reduce the presence of non-specific DNA, thereby minimizing background levels. This results in a increased sensitivity and better enrichment. The reduced background makes it possible to use the ChIP-IT High Sensitivity Kit with as little as 1,000 cells per immunoprecipitation reaction for high abundance proteins and as little as 50,000 cells per immunoprecipitation reaction for low abundance proteins. In the experiment below, ChIP was performed using the Histone H3K4me3 antibody and chromatin from 1,000 to 1 million cells. Significant enrichment was still detectable at the positive control GAPDH promoter when using only 1,000 cells.
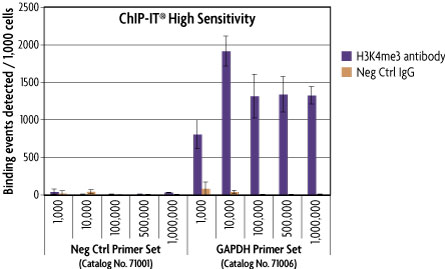
Figure 4: ChIP-IT High Sensitivity is suitable for use with limited sample material.
MCF-7 chromatin was prepared according to the ChIP-IT High Sensitivity manual using between 100,000 to 4 million cells per chromatin preparation. The immunoprecipitation was then performed using the indicated number of cells with Active Motif's Histone H3K4me3 antibody (Catalog No. 39915) and a negative control IgG. Following enrichment, qPCR was performed using the ChIP-IT qPCR Analysis Kit (Catalog No. 53029). The ChIP-IT High Sensitivity Kit shows strong enrichment with the positive control GAPDH promoter from as little as 1,000 cells and no enrichment with the negative control Untr12 primer set. Data is expressed as binding events detected per 1,000 cells which represents the average of the raw data triplicates adjusted for the amount of chromatin in the reaction, the resuspension volume and the primer efficiency.
Enriched DNA is ChIP-Seq Validated
The ChIP DNA obtained from the ChIP-IT High Sensitivity Kit is of high quality and can be used in downstream applications such as ChIP-Seq. Active Motif has validated the ChIP-IT High Sensitivity Kit for ChIP-Seq with both challenging antibodies that target low abundance proteins and highly abundant histone proteins as shown below.
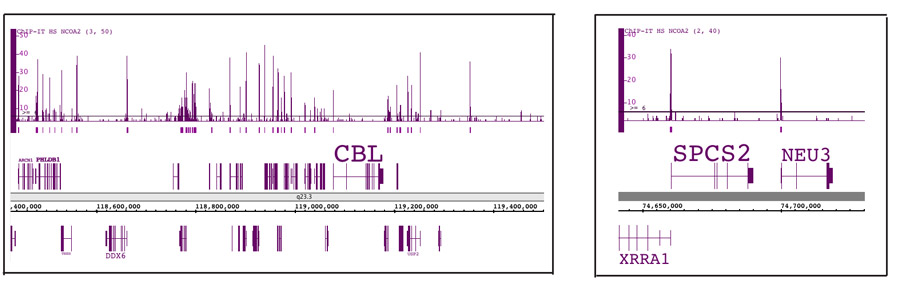
Figure 5: ChIP-Seq data for the low abundance transcription factor NCOA2.
ChIP was performed using the ChIP-IT High Sensitivity Kit, an antibody against the low abundance transcriptional co-activator NCOA2 and chromatin from 3 million MCF-7 cells. ChIP DNA was processed and 30 million sequence tags were generated using the Illumina® Next-Gen sequencing platform. The sequence data was analyzed to identify all NCOA2 binding sites in the human genome. The left panel shows multiple binding sites across a 1 million base pair region on chromosome 11. The image in the right panel is zoomed in to show NCOA2 peaks in the promoters of two genes.
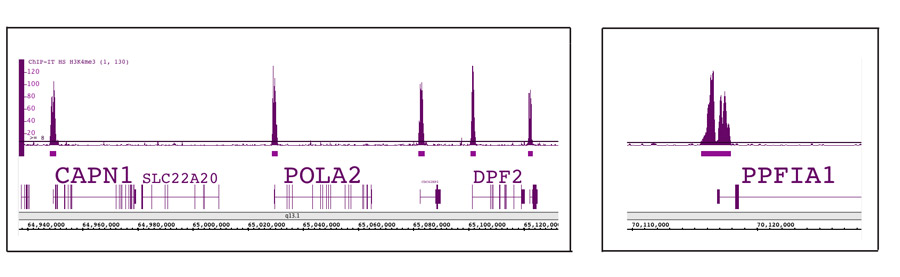
Figure 6: ChIP-Seq data for Histone H3K4me3.
ChIP was performed using the ChIP-IT High Sensitivity Kit, an antibody against Histone H3K4me3 (Catalog No. 39915) and chromatin from 1 million MCF-7 cells. ChIP data was processed and 24 million sequence tags were generated using the Illumina® Next-Gen sequencing platform. The sequencing data was analyzed to identify all the H3K4me3 binding sites in the human genome. The left panel shows the presence of the H3K4me3 in the promoters of 5 genes across a 175,000 bp region on chromosome 11. The image in the right panel is zoomed in to show binding in the promoter of a single gene. Binding at the PPFIA1 gene shows the common double peak with depletion right at the transcriptional start site.
FAQs
ChIP-IT High Sensitivity® FAQs
What’s the minimum number of cells required for the chromatin prep?
The ChIP-IT High Sensitivity Kit requires at least 100,000 cells for the chromatin prep, but this chromatin can then be aliquoted for multiple ChIP reactions.
What is the minimum and maximum amount of input chromatin I can use for the IP?
For antibodies against a high-abundance histone mark, this kit can achieve sensitive enrichment from chromatin from as little as 1000 cells (6.7 ng chromatin) per ChIP. For antibodies against transcription factors, we recommend using chromatin from 50,000 cells (330 ng) or more per ChIP.
For an upper limit, we recommend using no more than 30 𝝻g of chromatin, roughly the amount obtained from 4.5 million cells (using a rough estimate that 1.5 million cells yield 10 𝝻g chromatin). The ideal range of chromatin per IP for this kit is 10-30 𝝻g, the amount of chromatin from 1.5-4.5 million cells.
If I am working with suspension cells how should I fix the cells?
The protocol should be similar to adherent cells, starting with adding 1/10th volume Complete Cell Fixative solution directly to the cells in the culture media.
At what points can I stop in the protocol?
The protocol may be stopped at the following times and temperatures:
- After formaldehyde fixation and centrifugation (intact cell pellet), -80°C.
- After chromatin shearing, -80°C.
- After DNA clean-up, -20°C.
How do I shear my chromatin?
The ChIP-IT High Sensitivity Kit protocol has been validated using our EpiShear™ Probe Sonicator. For other sonicators such as Active Motif’s PIXUL™ Multi-Sample Sonicator or those by Diagenode and Covaris, shearing guidelines will vary. Please refer to the Tech Note: Sonication Recommendations for ChIP or contact Technical Support for further guidance.
I ran my sheared DNA from the ChIP-IT High Sensitivity Kit on a Bioanalyzer and it appears as if it is high molecular weight. Is the sonication not working?
The sonication may have worked correctly, but the ChIP-IT High Sensitivity buffers can cause artifacts to appear as higher molecular weight on a Bioanalyzer. We recommend the use of an agarose gel to analyze the chromatin shearing efficiency. Note that the kit manual includes specific recommendations for a high salt, high temperature denaturation step prior to visualization on the agarose gel. This is critical for proper visualization.
What antibodies can I use?
It is important to use a ChIP-validated antibody because SDS conditions used in IP can reduce antibody-protein binding. Many antibodies that perform well in other applications (e.g., Western) do not perform well in ChIP. Please see our list of ChIP-Validated Antibodies. If a ChIP validated antibody is not available, we suggest checking the literature for an antibody that has been used on crosslinked material (ex. Immunofluorescence (IF) or immunohistochemistry (IHC)).
Do you have a suggestion for improving cell lysis?
- If a dounce homogenizer is not enough to lyse cells effectively, try using a 28-30 gauge needle to lyse the cells. Draw the cells through the needle 4-5 times.
- Try reducing the fixation time.
- For especially difficult to lyse cells such as PBMCs (lymphocytes, monocytes and dendritic cells) use the High Sensitivity Chromatin Preparation Kit (53046) which contains a hypotonic Swelling Buffer for better lysis or switch to the ChIP-IT PBMC Kit (53042) for a complete ChIP solution for these cell types.
What controls should I use in my ChIP assay?
We suggest using qPCR primers specific for genomic regions where the factor or histone modification is known to be present (positive control) and where it’s known to be absent (negative control).
Additionally, one can include positive and negative antibody controls. A positive antibody control is one that would recognize proteins bound to the locus of interest. Some common positive antibody controls are Histone H3 or RNA Pol II. A negative antibody control would be one that does not recognize proteins bound to the locus of interest. A commonly used negative control antibody is mouse IgG.
For ChIP-seq, an “input DNA control” is normally used in order to identify false “peaks” or reveal regions of the genome that have been duplicated. This control has been fixed and sonicated but not immunoprecipitated. We suggest using 1-10% of the quantity of DNA that went into each ChIP reaction.
For convenience, Active Motif offers a number of accessory products containing controls to validate your ChIP experiments including ChIP-IT Control Kits, ChIP-IT qPCR Primer Sets, and a ChIP-IT qPCR Analysis Kit.
I’m running the ChIP-IT High Sensitivity Kit for the first time and I need to optimize the fixation, shearing, and lysis conditions. Is there any way to do some testing without using all of the chromatin prep reagents before moving forward with the immunoprecipitation step?
For optimizing the shearing, we offer our High Sensitivity Chromatin Prep kit. This companion module to our ChIP-IT High Sensitivity® Kit contains extra reagents specifically for optimizing the fixation, shearing, and lysis of the sample before proceeding to the full scale ChIP reaction.
Documents
ChIP-IT High Sensitivity® Documents