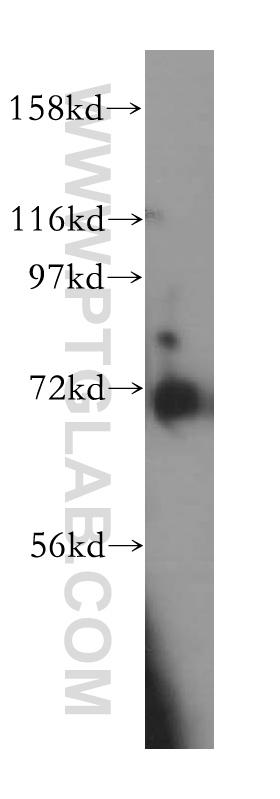
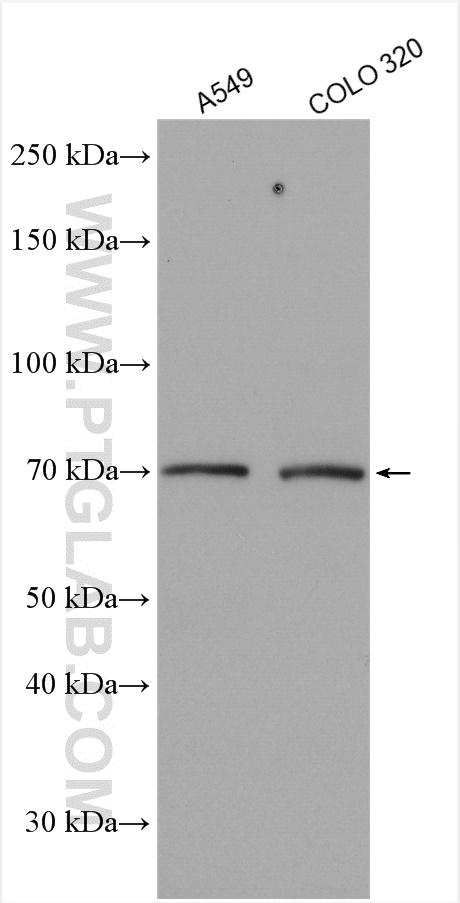
验证数据展示
经过测试的应用
| Positive WB detected in | COLO 320 cells, A549 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
13871-1-AP targets KBTBD2 in WB, ELISA applications and shows reactivity with human, mouse, rat samples.
| 经测试应用 | WB, ELISA Application Description |
| 经测试反应性 | human, mouse, rat |
| 免疫原 | KBTBD2 fusion protein Ag4823 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | kelch repeat and BTB (POZ) domain containing 2 |
| 别名 | BKLHD1, KBTBD2, KIAA1489 |
| 计算分子量 | 622 aa, 71 kDa |
| 观测分子量 | 60-70 kDa |
| GenBank蛋白编号 | BC047107 |
| 基因名称 | KBTBD2 |
| Gene ID (NCBI) | 25948 |
| RRID | AB_2296330 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | Q8IY47 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
Kelch repeat and BTB domain containing 2(KBTBD2), also known as BTB and kelch domain-containing protein 1, encodes a BTB-Kelch family subunit that p85α-specific recognition for the Cullin3-based E3 ubiqitin ligase complex, which regulates p85α protein ubiquitination and subsequently regulates phosphoinositide-3 kinases (PI3K) signaling (PMID:27708159). However, the expression profile and functional significance of Kbtbd2 is still largely unknown. The Human Protein Atlas which is an online source shows high Kbtbd2 expression in the spleen and thymus, suggesting that Kbtbd2 might involved in immunization (PMID:29331106). In addition, Kbtbd2 is related to Src-induced phosphorylation of protein, spermatogonial stem cell activity(PMID:15623525, 29331106). A 60-70 kDa band has also been reported(PMID:27708159).
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for KBTBD2 antibody 13871-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |