Methylated DNA Immunopreciptation (MeDIP) Kit
Methylated-DNA-Immunopreciptation-MeDIP-Kit-AM736
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
MeDIP Kit Overview
The MeDIP Kit is designed to immunoprecipitate and enrich single-stranded DNA (ssDNA) fragments containing 5-methylcytosine (5-mC). The MeDIP Kit contains a monoclonal 5-methylcytosine antibody, a bridging antibody, and the necessary buffers to perform methylated DNA immunoprecipitation. The high specificity and affinity of the antibody enables separation of 5-mC DNA from DNA containing 5-hmC and other DNA methylation variants using single-stranded input DNA.
Active Motif's fast, magnetic protocol has been streamlined to minimize the number of wash and incubation steps, saving you valuable time. The kit also includes MseI-digested human genomic DNA and PCR primers that can be used to verify the efficiency of the enrichment. Additionally, the kit contains a bar magnet for easy separation and elution of the enriched methylated DNA.
5-Methylcytosine (5-mC) Methylated DNA Immunoprecipitation
Methylated DNA Immunoprecipitation is an immunocapture technique in which an antibody specific for methylated cytosines is used to immunoprecipitate methylated genomic DNA fragments. MeDIP uses single-stranded DNA that has been fragmented by sonication or enzymatic digestion. The addition of a monoclonal antibody specific for 5-methylcytosine in the presence of a bridging antibody enables specific capture of the DNA fragments containing 5-mC methylation. The use of protein G magnetic beads provides quick and efficient capture. Methylated fragments are then eluted from the magnetic beads, resulting in DNA samples that are specifically enriched for 5-mC DNA methylation. This selectivity is important as most common approaches to analyze DNA methylation, such as enzymatic approaches and bisulfite conversion, are unable to distinguish between 5-mC and 5-hmC.
The high affinity of the 5-mC antibody used in the MeDIP Kit enables detection of methylated cytosines regardless of their context. This means that the methylation does not need to occur in the context of a CpG dinucleotide, unlike methyl-binding protein (MBD) affinity enrichment methods which requires CpG methylation for proper detection.
MeDIP Kit Advantages
- Specific antibody only enriches 5-mC methylated DNA
- Includes bridging antibody to increase efficiency of DNA capture
- One-step IP reaction
- No need to pre-wash magnetic beads
- Optimized magnetic protocol to minimize hands-on time
- Kit includes positive control human genomic DNA and PCR primers to verify efficiency
Flow Chart of MeDIP Method
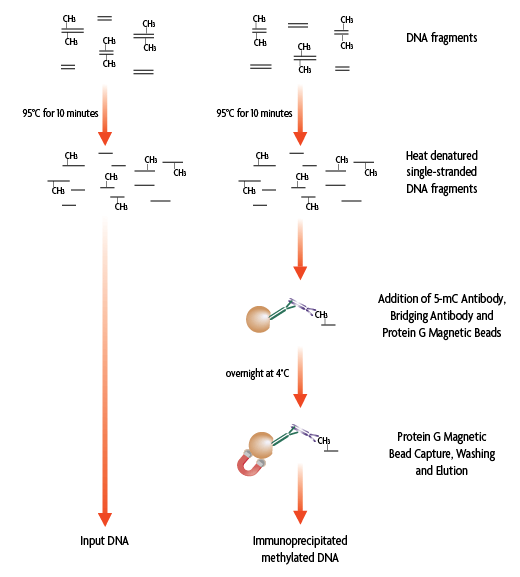
Figure 1: Flow chart of the MeDIP Method. Single-stranded, fragmented genomic DNA containing 5-methylcytosine can be specifically captured from the rest of the genomic DNA with the monoclonal 5-mC antibody in the presence of a bridging antibody and magnetic beads. Following an overnight incubation, the enriched DNA is then eluted from the beads.
Preparation of MeDIP-Enriched Samples for Next-Gen Sequencing (NGS)
Since the MeDIP Kit uses a 5-methylcytosine antibody that only binds methylated cytosines in the context of single-stranded DNA to enrich for methylated genomic DNA, the DNA must be denatured prior to immunoprecipitation. As a result of denaturation, the enriched DNA can not be processed for next-generation sequencing using some common NGS library generation protocols that include adapter ligation to double-stranded DNA.
This problem can be circumvented by either addition of the NGS adapters to fragmented genomic DNA prior to performing MeDIP or using a library preparation protocol that is compatible with ssDNA.
If you intend to perform NGS analysis following methylated DNA immunoprecipitation using the Active Motif MeDIP Kit, we suggest using the xGen ssDNA & Low-Input DNA LIbrary Preparation Kit from IDT following the manufacturer's instructions.
Contents
MeDIP Kit Contents
The MeDIP Kit contains a mouse monoclonal 5-methylcytosine antibody, a bridging antibody, negative control mouse IgG, PIC, Buffer C, Buffer D, Elution Buffer AM2, Neutralization Buffer, sterile water, PCR tubes, protein G magnetic beads, and a bar magnet for the separation and elution of DNA fragments containing 5-mC.
Protocols are provided for the fragmentation of sample DNA all the way through to real time PCR analysis of a locus of interest.
The kit also includes positive control DNA (MseI-digested human genomic DNA) and positive control ZC3H13 PCR primers.
Storage conditions range from room temperature to -20°C; please refer to the manual for proper storage temperatures of each component.
Data
MeDIP Kit Data
The MeDIP Kit is able to selectively enrich for DNA fragments containing 5-methylcytosine DNA methylation from the rest of the genomic DNA population. The MeDIP Kit was tested using 500 ng of the included MseI-digested human genomic DNA in the presence or absence of bridging antibody. Eluted DNA was purified with Active Motif's Chromatin IP DNA Purification Kit (Catalog No. 58002) and tested in real time PCR with the included ZC3H13 PCR primer set. Results shown in figure 2 demonstrate the importance of using a bridging antibody within the assay. In figure 3, enriched DNA was tested using either the included positive control ZC3H13 PCR primer mix or a negative control GAPDH PCR primer mix.
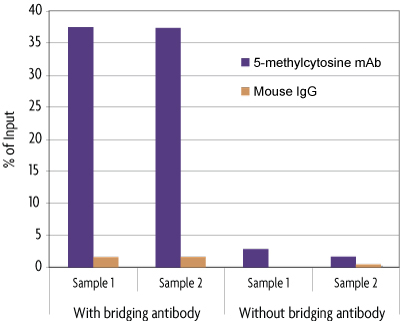
Figure 2: MeDIP Kit in the presence or absence of bridging antibody.
MseI-digested human genomic DNA (500 ng) was processed using the MeDIP Kit with the 5-methylcytidine mAb or negative control mouse IgG in the presence or absence of bridging antibody. Eluted DNA was purified and tested using real time PCR with the included ZC3H13 PCR primer mix. The 5-methylcytidine antibody specifically enriched methylated DNA in the presence of bridging antibody, but did not enrich for DNA with the mouse IgG or in the absence of bridging antibody. The ZC3H13 locus analyzed in this experiment is methylated in human genomic DNA and therefore should amplify. Data shown are the results from samples assayed in duplicate.
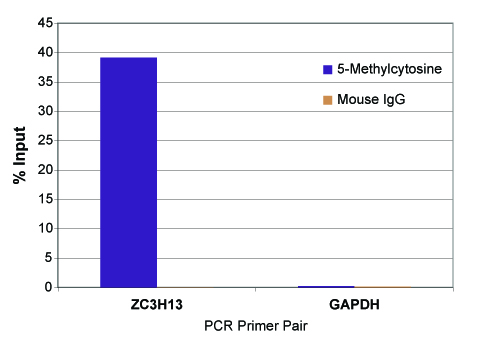
Figure 3: Real time PCR results of the control DNA with ZC3H13 and GAPDH PCR primer sets.
MseI-digested human genomic DNA (500 ng) was processed using the MeDIP Kit with the 5-methylcytosine mAb or negative control mouse IgG. Eluted DNA was column purified with Active Motif's Chromatin IP DNA Purification Kit (Catalog No. 58002) and tested using real time PCR with the included ZC3H13 PCR primer mix and a negative control GAPDH PCR primer mix. The 5-methylcytosine antibody specifically enriched methylated DNA with the ZC3H13 locus but did not enrich for DNA with either the GAPDH locus or the mouse IgG. Data shown are the results from samples assayed in duplicate.
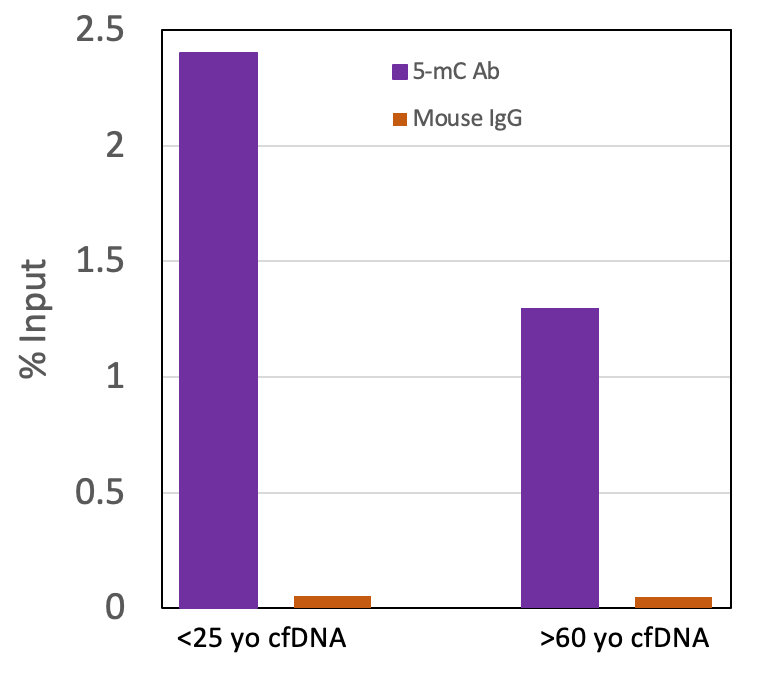
Figure 4: Active Motif’s MeDIP Kit can be used to enrich for methylation in cfDNA. Serum samples were collected in serum separator tubes (SST) from two healthy “normal” adults, one <25 yo and one >60 yo. Cell-Free DNA (cfDNA) was purified with the Active Motif Cell-Free DNA Purification Kit (Cat no. 25503/25504) and quantified with a Qubit fluorometer. Undigested cfDNA (200 ng) was processed using the Active Motif MeDIP Kit (Cat no. 55009) with the 5-mC Ab or negative control mouse IgG Ab in the kit. Eluted DNA was purified and tested using real time PCR with the included ZC3H13 PCR primer set. The 5-methylcytidine antibody specifically enriched methylated DNA but did not enrich for DNA with the mouse IgG. The ZC3H13 locus analyzed in this experiment is methylated in human genomic DNA and therefore should amplify.
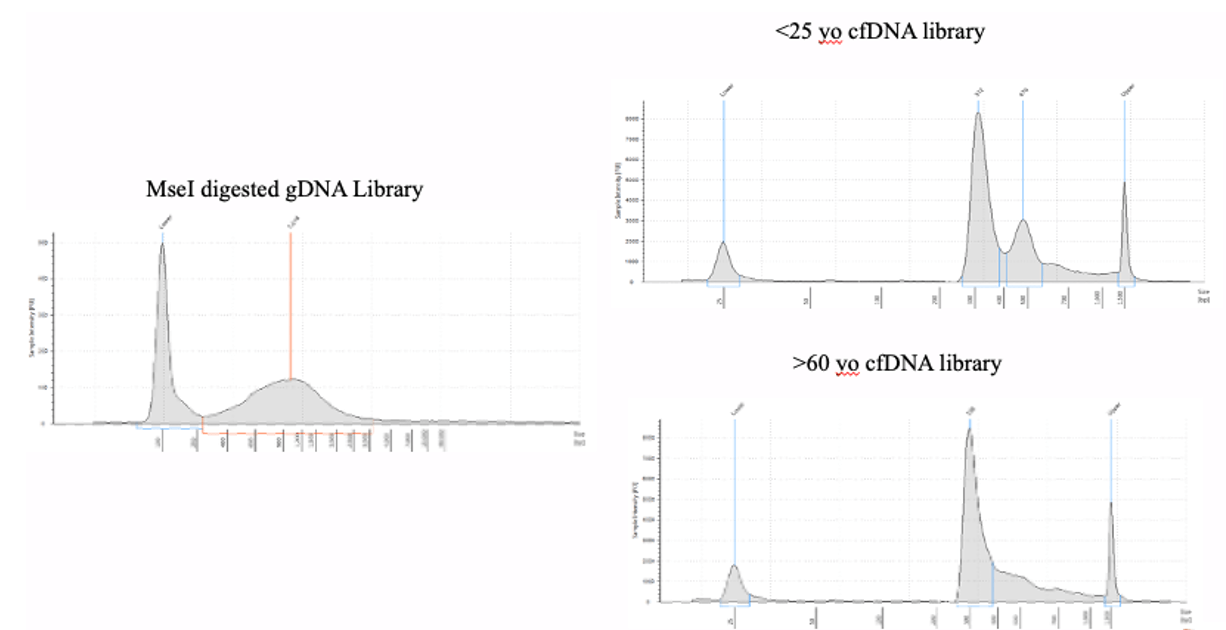
Figure 5: Example DNA Libraries from the Active Motif MeDIP Kit.
The Swift Biosciences Accel-NGS™ 1S PLUS DNA Library Kit, now the IDT xGen ssDNA & Low-Input DNA Library Preparation Kit (Cat# 10009859), was used to prepare libraries after using the Active Motife MeDIP Kit with MseI digested gDNA and undigested cfDNA.
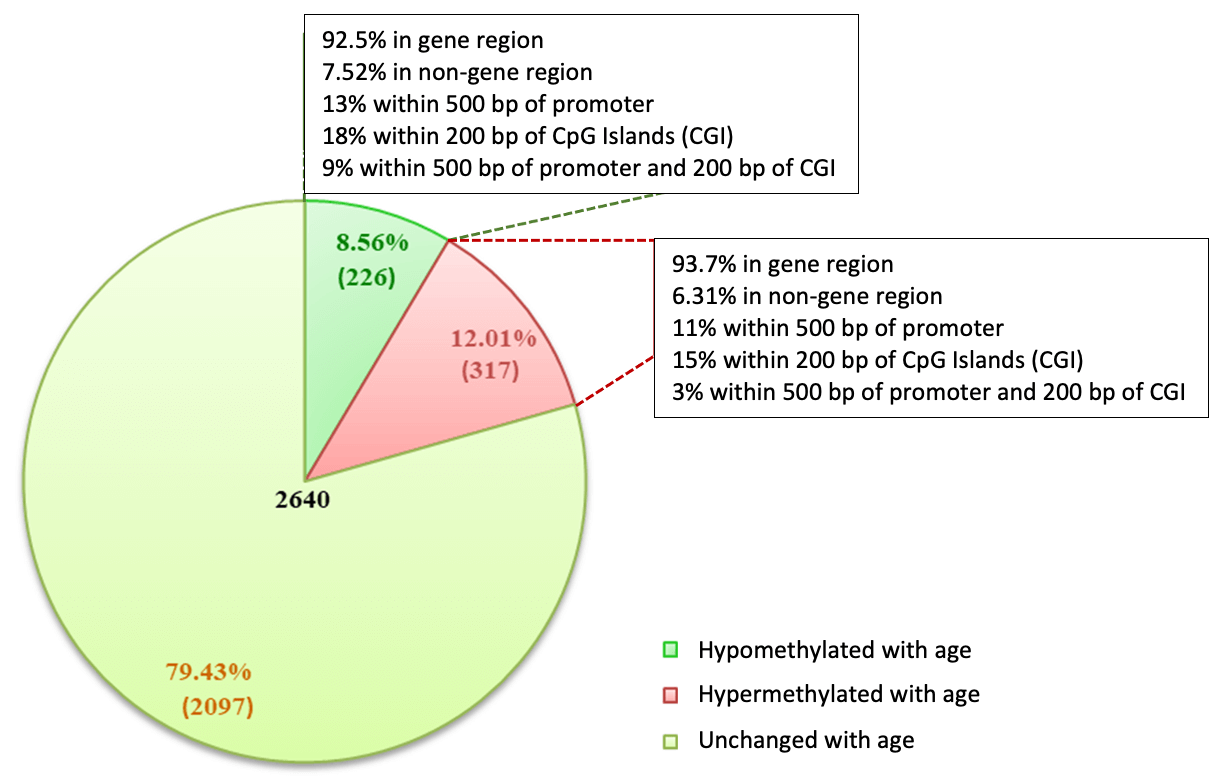
Figure 6: MeDIP-Seq uncovers aging-associated differential methylation patterns in cfDNA.
Serum samples were collected in serum separator tubes (SST) from healthy “normal” adults, two <25 yo and two >60. Cell-Free DNA (cfDNA) was purified with the Active Motif Cell-Free DNA Purification Kit (Cat no. 25503/25504) and quantified with a Qubit fluorometer. 200 ng cfDNA was used in the MeDIP Kit and libraries were prepared using the Swift Biosciences Accel-NGS™ 1S PLUS DNA Library Kit, now the IDT xGen ssDNA & Low-Input DNA Library Preparation Kit (Cat# 10009859). Input to library prep was determined using AluI quantification. MeDIP and Input libraries were sequenced (38bp PE chemistry on Next-Seq500) and processed as per Active Motif’s standard pipeline for MeDIP-Seq. Peaks called using MACS2 relative to input (p-value cutoff = 1.00x10-7).
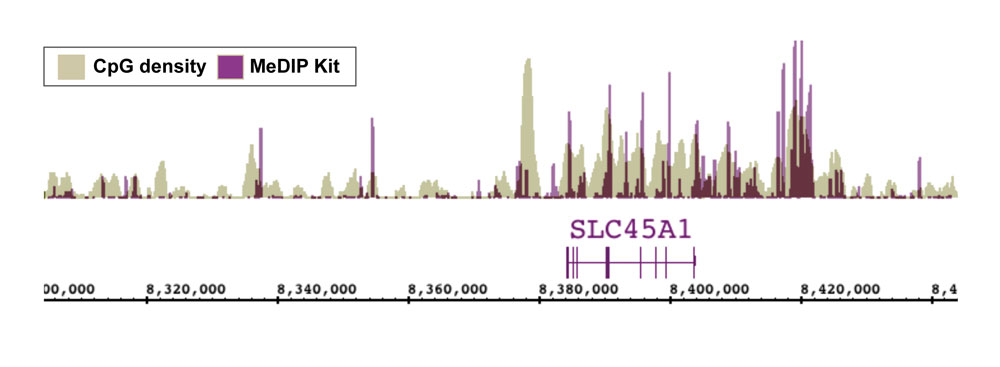
Figure 7: NGS data generated using Active Motif’s MeDIP Kit correlates well with CpG density.
DNA was enriched from 1 ug of adaptor-ligated human PBMC DNA using Active Motif’s MeDIP Kit. NGS was performed using the Illumina platform to generate 26 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows that the enriched regions (purple peaks) correlate well with CpG density (tan peaks).
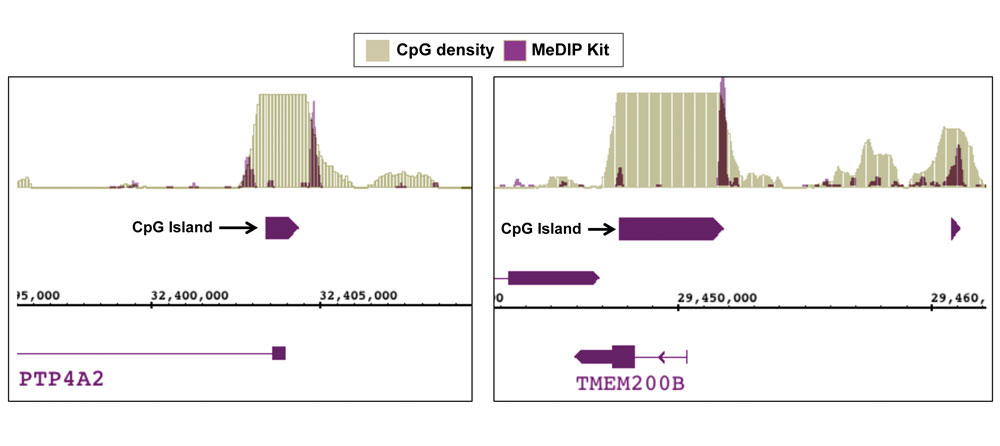
Figure 8: NGS data generated using Active Motif’s MeDIP Kit detects methylation at CpG shores.
DNA was enriched from 1 ug of adaptor-ligated human PBMC DNA using Active Motif’s MeDIP Kit. NGS was performed using the Illumina platform to generate 26 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows two examples of DNA methylation being detected at CpG shores rather than in the CpG island itself. This data agrees with recent findings showing that a large portion of methylated sites occur in these regions that are adjacent to CpG islands.
Documents
MeDIP Kit Documents
