验证数据展示
经过测试的应用
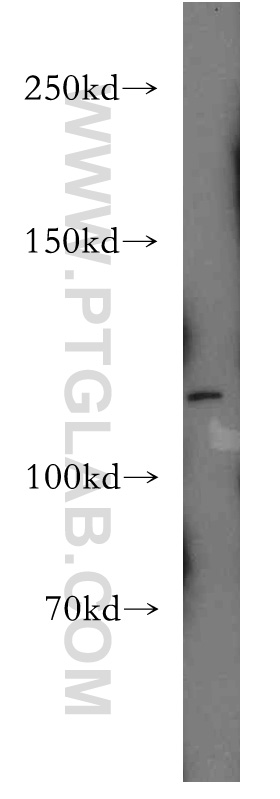
| Positive WB detected in | HeLa cells |
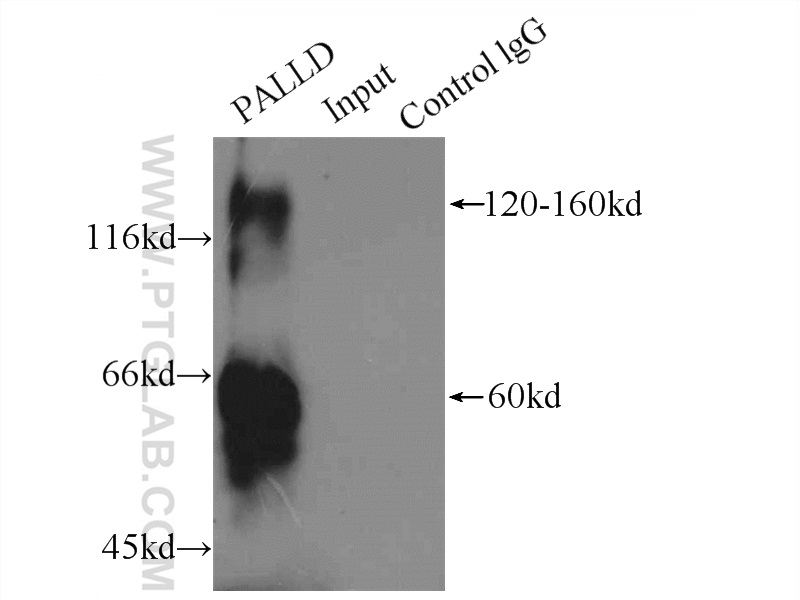
| Positive IP detected in | HeLa cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
16179-1-AP targets Palladin-Specific in WB, IP, ELISA applications and shows reactivity with human samples.
| 经测试应用 | WB, IP, ELISA Application Description |
| 经测试反应性 | human |
| 免疫原 | Peptide 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | palladin, cytoskeletal associated protein |
| 别名 | CGI 151, KIAA0992, palladin, PALLD, PNCA1, Sarcoma antigen NY SAR 77, SIH002 |
| 计算分子量 | 151 kDa |
| 观测分子量 | 120 kDa |
| GenBank蛋白编号 | NM_016081 |
| 基因名称 | palladin |
| Gene ID (NCBI) | 23022 |
| RRID | AB_10695626 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | Q8WX93 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
Palladin is a widely expressed protein found in stress fibers, focal adhesions, growth cones, Z-discs, and other actin-based subcellular structures. It belongs to a small gene family that includes the Z-disc proteins myopalladin and myotilin. It is a cytoskeletal protein required for organization of normal actin cytoskeleton. It functions in establishing cell morphology, motility, cell adhesion and cell-extracellular matrix interactions in a variety of cell types. This antibody, generated against N-terminal peptide of palladin, recognizes 200 kDa, 140 kDa, and 65 kDa isoforms of this protein.
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for Palladin-Specific antibody 16179-1-AP | Download protocol |
| IP protocol for Palladin-Specific antibody 16179-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |