RNA Subcellular Isolation Kit
RNA-Subcellular-Isolation-Kit-AM1160
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
The RNA Subcellular Isolation Kit provides an efficient method to isolate nuclear and cytoplasmic RNA without cross-contamination. This method can be used to isolate RNA sequences as small as 75 nucleotides (e.g. snoRNA, mRNA, and lncRNA). RNA molecules exist at different levels of maturation and processing within a cell at any given time. Such heterogeneity can bias downstream analysis and interpretation of data. By isolating RNA from specific subcellular fractions, the accuracy of gene expression analysis can be greatly improved. Subcellular isolation enhances the ability to detect low abundance transcripts and aids in the ability to study RNA processing dynamics by reducing the background from other intronic or mature RNAs. Additionally, subcellular isolation helps to localize long non-coding RNAs (lncRNAs) to their site of action, which can provide important insights into their functional role.
The RNA Subcellular Isolation Kit contains enough reagents to perform 15 nuclear and 15 cytoplasmic purifications from 2-4 million cultured cells or 15 mg of tissue per reaction. Simply lyse your samples, then separate the nuclear and cytoplasmic RNA by centrifugation. Each RNA fraction is loaded onto individual spin columns and washed to remove any proteins. Finally, the purified RNA is eluted from the column and quantified. The assay can also be used to isolate total RNA.
To learn more about our RNA Subcellular Isolation Kit, click on the Description, Method or Contents tabs below. To view a manual or other related documents, click on the Documents tab below.
Description
Kit Specifications:
- Separates nuclear and cytoplasmic RNA without cross-contamination
- Works with cultured cells or tissue
- Easy-to-use spin columns included for RNA purification
- Method avoids the use of phenolic compounds
- Isolates RNA between the sizes of 75-7,000 nucleotides (includes snoRNA, mRNA, lncRNA)
- Purified RNA validated for use in RT-qPCR and RNA-seq
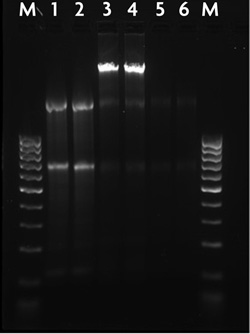
Figure 1: Agarose gel of nuclear and cytoplasmic RNA fractions.RNA was isolated as nuclear and cytoplasmic fractions using the RNA Subcellular Isolation Kit. Following purification, half of the nuclear RNA was treated with DNase I to remove genomic DNA contamination using the DNase I Treatment Kit (Active Motif Catalog No. 25502). 1 µg of each RNA fraction was loaded onto a 1.5% gel for analysis.
Lane M = 100 bp DNA ladder
Lanes 1-2 = Cytoplasmic RNA
Lanes 3-4 = Nuclear RNA before DNase I treatment
Lanes 5-6 = Nuclear RNA after DNase I treatment

Figure 2: TapeStation trace of cytoplasmic RNA isolated with the RNA Subcellular Isolation Kit.Cytoplasmic RNA was isolated from HeLa cells using the RNA Subcellular Isolation Kit. TapeStation analysis was performed to analyze the recovered RNA before and after rRNA depletion. The yellow and green traces represent two cytoplasmic samples pre-RNA depletion. The two large peaks represent the 18S and 28S rRNA. The blue and red traces represent the same samples after rRNA depletion. The 18S and 28S peaks are no longer present. The size of the purified RNA spans from 75-7000 nucleotides.

Figure 3: Small RNA distribution frequency between cytoplasmic and nuclear fractions.Cytoplasmic and nuclear RNA were isolated from HeLa cells using the RNA Subcellular Isolation Kit. RNA was processed with the NEBNext® Multiplex Small RNA Library Prep Kit and size selected to remove large RNA. 75 bp single read sequencing was performed and the data was mapped to miRBase. A frequency of the small RNA (sRNA) distribution for each fraction shows the specificity of subcellular isolation.
Applications:
The RNA Subcellular Isolation Kit is designed to isolate nuclear and cytoplasmic RNA from cultured cells and fresh tissue, or total RNA from cultured cells and fresh or frozen tissue. The purified RNA is suitable for use in a variety of downstream applications.
- Reverse-transcription quantitative PCR (RT-qPCR)
- RNA-seq
- Northern blotting
- Purified RNA can undergo DNase I treatment
- Purified RNA can undergo ribosomal RNA (rRNA) depletion
- Purified RNA can be used for polyadenylation (poly A) selection

Figure 4: Subcellular RNA Isolation to identify RNA localization.Nuclear, cytoplasmic and total RNA were isolated from HeLa cells using the RNA Subcellular Isolation Kit. Following purification and DNase I treatment of the nuclear and total RNA fractions with Active Motif's DNase I Treatment Kit (Catalog No. 25502) purified RNA was subjected to reverse transcription and qPCR. Each subcellular fraction was plotted as a percentage of total RNA. The data shows the localization of various RNAs. MALAT1, NEAT1 and TUG1 are lncRNAs. Their expression data is consistent with published RNA-FISH data for the same RNAs and cell type. Both S14, which is a ribosomal RNA, and BIRC5, a mRNA, are expected to localize within the cytoplasm as demonstrated above.

Figure 5: RNA-Seq data confirms lncRNA localization.Nuclear, cytoplasmic and total RNA were isolated from HeLa cells using the RNA Subcellular Isolation Kit. RNA was subjected to ribosomal RNA depletion during NGS library preparation. Samples were then sequenced using the Illumina HiSeq and 100 bp paired end reads with 50 M reads per sample. NEAT1 is a lncRNA that is primarily located in the nucleus, while TUG1 is a lncRNA known to have both nuclear and cytoplasmic localizations (see Figure 3 above for RT-qPCR data). This data is also consistent with published RNA-FISH data and RT-qPCR analysis.
Method
How does it work?
The RNA Subcellular Isolation assay works by lysing cell or tissue samples in complete lysis buffer. The lysate is then separated by centrifugation, with the supernatant containing cytoplasmic RNA and the pellet containing nuclear RNA. A guanidine-based buffer and ethanol are added to each RNA fraction before being loaded onto individual spin columns. Following a wash step to remove any proteins, the RNA fractions are eluted and quantified. RNA can be further process with Active Motif's DNase I Treatment Kit (Catalog No. 25502) to remove any genomic DNA contamination from the purified RNA sample prior to downstream analysis.
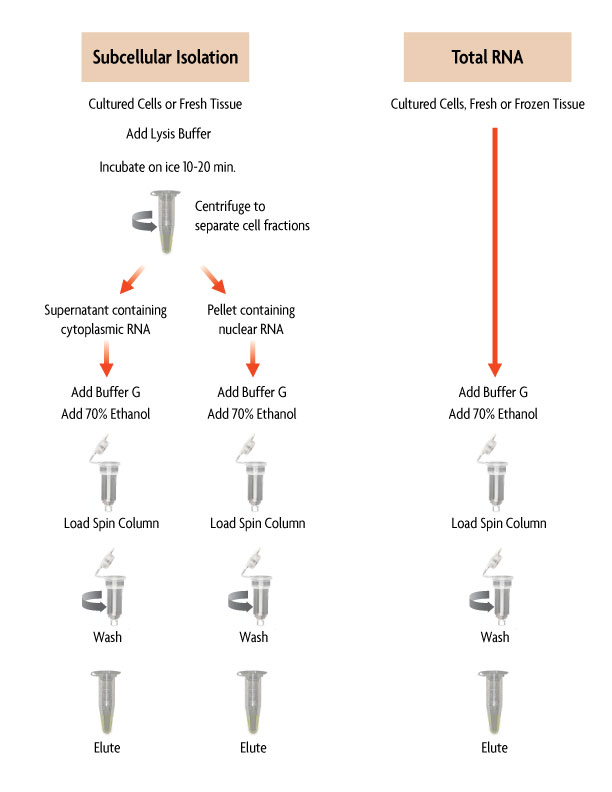
Figure 1: Flow Chart of the RNA Subcellular Isolation Method.
| Maximum Column Binding Capacity | 40 µg |
| Maximum Column Loading Volume | 750 µl |
| Size of purified RNA | 75-7,000 nucleotides |
| Maximum Amount of Starting Material Cells Tissue |
2 x 106 cells (Total RNA) 4 x 106 cells (Cytoplasmic and Nuclear RNA) 15 mg tissue |
| Assay Time | 2 hours |
| Average Yields HeLa Cytoplasmic RNA HeLa Nuclear RNA HeLa Total RNA |
12-25 µg at 250-500 ng/µl from 2 x106 cells 12-25 µg at 250-500 ng/µl from 2 x106 cells 25-40 µg at 500-900 ng/µl from 2 x106 cells |
Contents
Contents & Storage
Please note that the RNA Subcellular Isolation Kits are shipped on dry ice and contain reagents with multiple storage temperatures inside. Please store each component at the temperature indicated below. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.This kit includes the following components:
- 1 M DTT; Store at -20°C
- 10X PBS; Store at 4°C
- Lysis Buffer AM7; Store at 4°C
- Buffer G; Store at 4°C
- Purification Columns; Store at RT
- DEPC Water; Store at RT
