TransAM® STAT3 & STAT Family Kits
TransAM-STAT3-STAT-Family-Kits-AM232
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
TransAM® Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue and cell extracts. Assays are available for over 40 different targets (see the list at right). Each kit includes a 96-stripwell plate in which multiple copies of a specific double-stranded oligonucleotide have been immobilized. When nuclear or whole-cell extract is added, activated transcription factor of interest binds the oligonucleotide at its consensus binding site and is quantified using the included antibody, which is specific for the bound, active form of the transcription factor being studied. For complete details, click the TransAM® Method tab below.
TransAM STAT Kits provide everything needed to study Signal Transducers and Activators of Transcription (STAT), including a positive control extract. TransAM STAT3 Kits are available with antibodies specific for the activated form of STAT3. A TransAM STAT Family kit is also available with the ability to independently screen the members of the STAT family (STAT1α, STAT3, STAT5A & STAT5B) simultaneously. The STAT1α, 5A and 5B antibodies can be used with human and rat extracts, while the STAT3 antibodies can be used with human, mouse and rat extracts. See the STAT Info tab below for kit data and more information; the kit manual can be downloaded under the Documents tab.
"Overall, I’ve been very pleased with these kits, particularly for STAT3 and C/EBP-beta. They are sensitive, easy/straightforward to use, and ultimately have given me incredibly interesting and valuable data for my research."
Darius Mcphail, PhD Student
Cardiff University
Documents (8)
STAT Info
STAT Transcription Factor Info
Signal Transducers and Activators of Transcription (STAT) are a family of latent cytoplasmic proteins that are activated in response to cytokines and growth factors. The STAT family has seven members: STAT1, 2, 3, 4, 5A, 5B and 6. STATs are involved in many pathways that regulate cellular functions in the lympho-hematopoietic system. STATs can be activated by ligands such as interferons (IFNs), epidermal growth factor (EGF), platelet derived growth factor (PDGF) and interleukins (IL). When members of the STAT family are phosphorylated they form homo- or heterodimers and translocate to the nucleus to activate transcription. STAT1α homodimerizes upon tyrosine or serine phosphorylation induced by IFNγ, but forms heterodimers with STAT2 when induced by IFNα/β. STAT3 is activated by phopshorylation of tyrosine 705 and serine 727 to mediate the expression of genes involved in cell growth and apoptosis. STAT5A and STAT5B are activated in response to several cell ligands and growth hormones. They are capable of serving as both transcriptional activators and signal transducers.
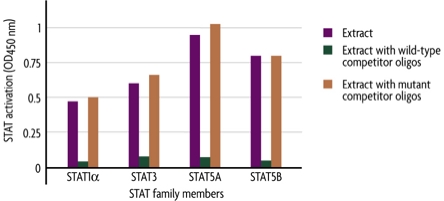
Figure 1: Monitoring STAT Family member activation using the TransAM STAT Family Kit.
STAT1α, STAT3, STAT5A and STAT5B activation were assayed using the TransAM STAT Family Kit. 1:1000 dilutions of each antibody in the kit were tested using 5 µg/well of nuclear extract prepared from a stimulated cell line: STAT1α was tested with U-937 (TPA + IFNγ), STAT3 with Hep G2 (IL-6, 100 ng/ml), and STAT5A and STAT5B with Nb2 (prolactin). Assays were performed in the absence or presence of 20 pmol of competitor oligonucleotide that contains either a wild-type or mutated STAT consensus binding site. Note that the wild-type oligonucleotide reduces STAT binding by over 90%, while incubation with the mutant STAT competitor oligo has a limited effect on STAT 1α, STAT3, STAT5A or STAT5B binding to DNA.
TransAM® Method
The TransAM® transcription factor ELISA advantage
Historically, transcription factor studies have been conducted using gelshift, Western blot and reporter plasmid transfections, which are time-consuming, do not allow for high-throughput and provide only semi-quantitative results. TransAM assays are up to 100 times more sensitive than gelshift techniques, and can be completed in less than 5 hours. Because TransAM is an ELISA-based assay*, there is no radioactivity, and the high-throughput stripwell format enables simultaneous screening of 1-96 samples. Inconsistencies due to variable reporter plasmid transfections are eliminated, along with the need to construct stable cell lines.
Why use TransAM® transcription factor ELISAs?
- Up to 100-fold more sensitive than gelshift assays
- Eliminates the use of radioactivity and the need to run gels
- Results in less than five hours
- Colorimetric readout enables easy, quantitative analysis with spectrophotometry at 450 nm
- 96-stripwell format enables both high and low throughput
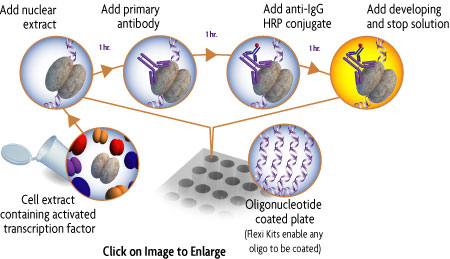
How TransAM® transcription factor ELISAs work
The TransAM format is perfect for assaying transcription factor binding to a consensus-binding site. TransAM Kits contain a 96-stripwell plate to which the consensus-binding site oligo has been immobilized. Activated nuclear extract is added to each well and the transcription factor of interest binds specifically to this bound oligonucleotide. A primary antibody specific for an epitope on the bound and active form of the transcription factor is then added followed by subsequent incubation with secondary antibody and Developing Solution to provide an easily quantified, sensitive colorimetric readout (Figure 1).
Figure 1: Flow chart of the TransAM process.
Activated transcription factor in the cell extract binds to the consensus-binding site on the oligo immobilized in the well. Incubation with the supplied primary and secondary antibodies specifically quantifies the amount of activated transcription factor.
* Technology covered under EAT-filed patents and licensed to Active Motif. Use of TransAM in NFκB-related drug discovery may be covered under U.S. Patent No. 6,150,090 and require a license from Ariad Pharmaceuticals (Cambridge, MA, USA).
Contents
Contents & Storage
One, two or five STAT 96-well assay plate(s) with plate sealer(s), STAT3 or STAT1α, STAT3, STAT5A and STAT5B primary antibodies, HRP-conjugated secondary antibody, STAT wild-type and mutated oligonucleotides, positive control cell extract, DTT, Protease Inhibitor Cocktail, Lysis, Binding, 10X Washing and 10X Antibody Binding Buffers, and Developing and Stop Solutions. Reagent storage conditions vary from room temperature to -80°C, see manual for details. All reagents are guaranteed stable for 6 months when stored properly.
TransAM® FAQs
Can I use your TransAM kit with my species of sample?
All of the TransAM kits will work with human samples. Some have cross-reactivity with mouse or rat. Each TransAM manual contains cross-reactivity information under the Kit Performance and Benefits section.
What is the recommended developing time for the colorimetric TransAM assays?
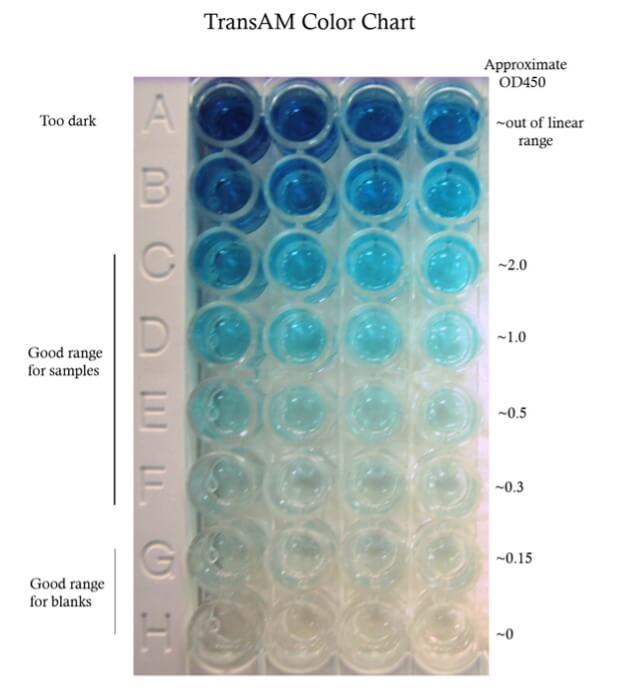
A recommended development time is given in the technical data sheet (TDS) for each lot. However, this time is only a guideline. It is important to monitor the color development for your particular samples. The TransAM color chart gives some guidelines for colorimetric assays.
What is the minimum amount of extract (and minimum number of cells) I can use per well?
Each of our specific TransAM kits has a different detection limit, which is indicated in each manual. For example, our TransAM NF-kB kits can detect binding from as little as 0.5 ug nuclear extract. Using a rough estimate yield of 20 ug nuclear extract from 1 million cells from our Nuclear Extract Kit, 0.5 ug nuclear extract can be obtained from approximately 25,000 cells.
How do I determine how much lysate to use per well?
The optimal protein concentration must be empirically determined. We recommend performing a small titration experiment– 5 ug, 10 ug, 20 ug for nuclear extract, more for whole cell extract, in order to ensure that the amount used per well is within detectable levels. You want to get both the treated and control sample signals within the linear range of the assay. See the TransAM color chart.
Can I increase the volume of sample added per well for the TransAM assay?
Yes, you can increase the total volume of sample added and scale up the reaction, but the ratio of Complete Binding Buffer to Complete Lysis Buffer must remain the same. Please note that there may not be enough reagents to perform all 96-rxns if you scale up and do not exceed a total volume of 100 µl. (Complete Binding Buffer plus sample diluted in Complete Lysis Buffer)
Why do I need to read the absorbance at both OD450nm and OD655nm?
The purpose of the reference wavelength (OD655nm) is to correct or normalize any differences in absorbance that are not due to the analyte. Everything being equal, the reference absorbance should not change between samples. But because of factors like variability in detection equipment, wells, etc. the absorbance can vary. Subtracting the reference from the specific reading: OD450nm - OD655nm accounts for these variations. Often the spectrophotometer or the program automatically does this.
Why do I need to run this assay in duplicates?
We recommend at a minimum running the assay in duplicate to get an idea of technical variability. If your assay has a high variability, we recommend doing triplicates. Plot the data with standard error to get a measure of the variability.
Is a standard curve required?
A standard curve may be used to determine the concentration of the target in your samples. However, determining the exact concentration is not required. In fact, a relative comparison between treated and control samples can be more informative than just the absolute concentration. Another reason to utilize a standard curve is to determine the correlation between protein concentration and the detected signal and ensure that samples fall within the linear range. With the TransAM kits, this range has already been determined and as long as your signals read below 2.0 OD450, the samples are within the linear range of the microplate reader’s detectability.
How do I know if the standard curve I generated is reasonable?
The R² value of the linear regression line should be 0.94 or higher for the standard curve. To increase this value, perform more replicates and pipet with precision.
What is the difference between the colorimetric and the chemiluminescent versions of the TransAM kits?
Generally speaking, a chemiluminescent ELISA is more sensitive and offers a greater dynamic range as compared to colorimetric ELISAs.
What’s the difference between the TransAM family kits and the TransAM individual kits?
Our TransAM family kits come with two 96-well plates and antibodies against multiple transcription factors within a family. Each antibody is provided in enough abundance to cover one whole 96-well plate, so there is flexibility to assay two or more related antibodies. Our TransAM individual kits each come with one 96-well plate and enough of one antibody to cover that plate.
What is the TransAM NF-kB Flexi kit?
Not all NF-kB sites are the same and our Flexi kits allow users to design and bind their own custom DNA oligonucleotides to the plate. This allows for the study of NF-kB dimers which bind to specific endogenous DNA sequences.
How should I prepare my nuclear extract?
Any of the following are compatible.
- Active Motif’s Nuclear Extract Kit (Cat. No. 40010/40410)**
- Preparation of Nuclear Extract protocol in the Appendix of the TransAM manual.
- Your own nuclear extract protocol, as long as the lysis buffers do not contain SDS. Be sure to resuspend the nuclear pellet in Complete Lysis Buffer.
** Note that some TransAM kits contain Lysis Buffer AM1 while others contain Lysis Buffer AM2. If working with a TransAM kit containing Lysis Buffer AM1, use the Nuclear Extract Kit as written. If working with a TransAM kit containing Lysis Buffer AM2, prepare the Complete Lysis Buffer in the Nuclear Extract Kit using Lysis Buffer AM2 rather than the AM1 that comes in the kit. If you have already prepared your samples using the Lysis Buffer AM1 from the Nuclear Extract Kit, we recommend that you dilute these 1:3 with the Lysis Buffer AM2 that came with your TransAM assay.
The TransAM manual contains instructions for the preparation of nuclear extract from cultured cells. Do you have a protocol for tissue?
The Active Motif Nuclear Extract Kit (Cat. No. 40010/40410) contains a protocol for creating a nuclear extract from tissue. You can also use your own tissue nuclear extract protocol as long as you resuspend the nuclear pellet in the Complete Lysis Buffer recommended in the TransAM kit manual. Avoid using SDS as it can interfere with the assay.
For the nuclear extraction what should I see under the microscope when I monitor cell lysis?
Under a phase contrast microscope intact cells should appear as a dark central region (nucleus) surrounded by a halo of less dense cytoplasm. In lysed cells, the nuclei will appear as dots surrounded by asymmetric debris. See example below:
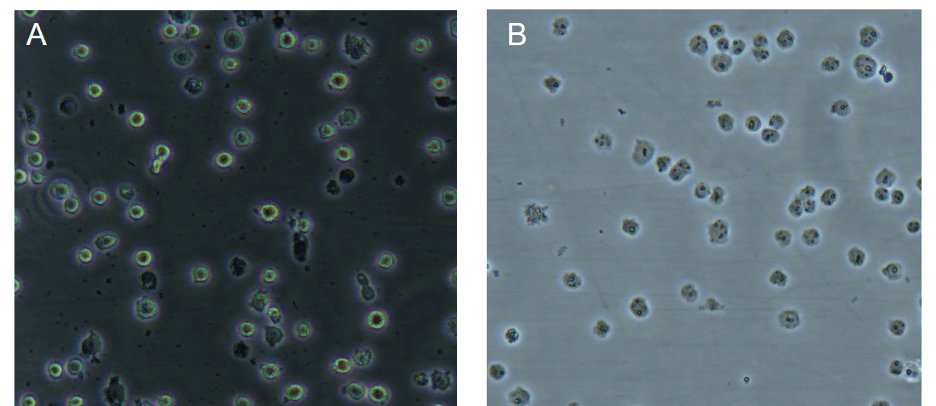
Figure: Images under phase contrast microscope of Jurkat cells before (A) and after (B) cell lysis; published with permission from Ziwei Wang, Tsinghua University.
Do you have any recommendations for nuclear extraction from PBMCs?
PBMCs are more difficult to lyse and can be resistant to lysis by the Hypotonic Buffer. Verify cell lysis efficiency using phase-contract microscopy. If cells are not adequately lysed, use a dounce homogenizer with a small clearance pestle (0.025–0.076 mm) to homogenize the sample or draw the sample through a 28-30 gauge needle several times for mechanical shearing. Active Motif’s Dounce Homogenizers for 1 mL capacity (Cat. No. 40401) and 15 mL capacity (Cat. No. 40415) both come with a small and large clearance pestle.
Do you have any recommendations for nuclear extraction from primary cells?
Primary cells can be challenging because they are typically fragile and can easily lyse with just scraping. Also, the number of cells is often limited. Be sure to monitor cell lysis under the microscope to maximize the extraction yield.
What dounce homogenizer should I use for the nuclear prep?
For cells, we recommend a dounce homogenizer with a small pestle clearance (0.025–0.076 mm) to lyse the cells and release the nuclei. For tissue we recommend a dounce homogenizer with a large pestle clearance (0.089–0.14 mm) to first homogenize the tissue into a single cell slurry and then a dounce homogenizer with a small pestle clearance to lyse the cells and release the nuclei. Active Motif’s Dounce Homogenizers for 1 mL capacity (Cat. No. 40401) and 15 mL capacity (Cat. No. 40415) both come with a small and large clearance pestle.

How many times can I freeze-thaw my nuclear extract?
It’s best to aliquot the extracts so that freeze-thaw cycles are not required. However, if you must thaw, we do not recommend freeze-thawing more than 2 times. Multiple freeze-thaws degrade proteins and destroy protein activity. Thaw the extracts on ice using pipetting to mix. Do you not vortex protein samples vigorously.
Can I use frozen samples to create the nuclear extracts?
We recommend using fresh cells or tissue, as the freezing process can disrupt the cytoplasmic and nuclear compartments, making it difficult to get pure fractions. However, if necessary, snap freeze tissues at -80°C. Cryopreserve cells before freezing at -80°C.
I am preparing a nuclear extract. The nuclear pellet is viscous and is not resuspending well in the Complete Lysis Buffer. How can I get it into solution?
To enhance the solubility of the pellet:
- Pipet up and down a few times with a large bore pipette tip.
- Use a dounce homogenizer.
- Vortex on high for 10 seconds.
- Increase the 30-minute incubation on the rocking platform to an hour.
If needed add 5-10 ul of Complete Lysis Buffer. Be sure not to overdilute the nuclear sample. Note that certain cell types are more likely to produce a viscous nuclear pellet and yet still give good protein yields, even if it doesn’t completely disappear. Do not add detergent as it could interfere with the TransAM assay.
Can I use whole cell extract?
It depends. With a whole-cell extract, the transcription factor of interest will be diluted into a larger volume than in a nuclear extract. So, more protein (2x or more) will be required. Also, if the transcription factor is regulated by nuclear-cytoplasmic localization, using whole-cell extract could affect the functional aspect of the assay.
Can I use the TransAM kit with cytoplasmic extracts?
It is not recommended for several reasons. The TransAM kit is designed to measure protein-DNA binding, which exclusively occurs in the nucleus. Any signal with the cytoplasmic extract may not be physiologically relevant. Also, the cytoplasmic extract is going to be much more dilute as compared to nuclear extract and will contain detergents unlike the nuclear fraction that that may interfere with the assay.
How should I quantify my nuclear extract protein?
We recommend using a Bradford Assay to quantify the protein rather than BCA. A BCA assay requires a higher dilution factor to eliminate confounding interfering agents and can limit the detection of protein in all but very concentrated samples. If using the Nuclear Extract Kit (Cat. No. 40010/40410), we recommend a dilution factor of 1:50 in water. There is a protocol for creating a BSA standard curve in the Appendix of the Nuclear Extract Kit manual.
How many cells/tissue should I use for the nuclear extract?
We suggest isolating nuclear extracts from samples of approximately 8.8 x106 cells or 45 mg tissue. A tissue conversion factor for general estimation is 2x105 cells/mg.
What protein yield can I expect from 8.8 x 106 cells?
This will depend on your cell or tissue type, but in general you should expect 150-250 µg at ~3-5 mg/ml from 8.8 x 106 cells.
What components can interfere with the TransAM assays?
SDS or sodium azide in your cell lysis buffer will inhibit the binding and can render your sample incompatible.