验证数据展示
经过测试的应用
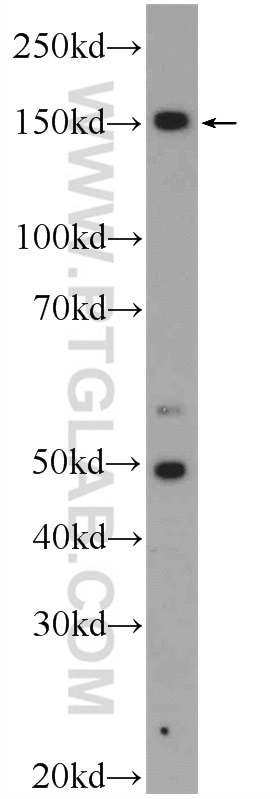
| Positive WB detected in | HEK-293 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
发表文章中的应用
| KD/KO | See 10 publications below |
| WB | See 16 publications below |
产品信息
23111-1-AP targets ZRANB3 in WB, ELISA applications and shows reactivity with human samples.
| 经测试应用 | WB, ELISA Application Description |
| 文献引用应用 | WB |
| 经测试反应性 | human |
| 文献引用反应性 | human |
| 免疫原 | ZRANB3 fusion protein Ag19407 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | zinc finger, RAN-binding domain containing 3 |
| 别名 | ZRANB3 |
| 计算分子量 | 1079 aa, 123 kDa |
| 观测分子量 | 150 kDa |
| GenBank蛋白编号 | BC064616 |
| 基因名称 | ZRANB3 |
| Gene ID (NCBI) | 84083 |
| RRID | AB_2744527 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | Q5FWF4 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
ZRANB3, also named Annealing helicase 2 or DNA annealing helicase and endonuclease ZRANB3, is a 1079 amino acid protein, which contains one RanBP2-type zinc finger, one helicase C-terminal domain, one HNH domain and one helicase ATP-binding domain. ZRANB3 localizes in the nucleus and belongs to the SNF2/RAD54 helicase family. ZRANB3 as a DNA annealing helicase and endonuclease is required to maintain genome stability at stalled or collapsed replication forks by facilitating fork restart and limiting inappropriate recombination that could occur during template switching events.
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for ZRANB3 antibody 23111-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |
发表文章
| Species | Application | Title |
|---|---|---|
Nat Commun Excessive reactive oxygen species induce transcription-dependent replication stress
| ||
Mol Cell Active DNA damage eviction by HLTF stimulates nucleotide excision repair.
| ||
Mol Cell Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.
| ||
Mol Cell Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops.
| ||
Mol Cell CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress.
| ||
Nat Commun Replication fork reversal triggers fork degradation in BRCA2-defective cells.
|