hMeDIP
hMeDIP-AM699
Synonyms
| Name | Format | Cat No. | Price | Delivery |
|---|
Overview
The hMeDIP Kit is designed to immunoprecipitate and enrich for DNA fragments containing 5-hydroxymethylcytosine (5-hmC). The hMeDIP Kit contains a highly specific purified 5-hmC antibody and the necessary buffers to perform methylated DNA immunoprecipitation (MeDIP). The affinity of the antibody enables separation of 5-hmC DNA from 5-mC DNA. Additionally, the antibody works to efficiently immunoprecipitate either double-stranded or single-stranded input DNA. For complete details, click the 5-hmC Info tab below.
Active Motif's fast, magnetic protocol has been streamlined to minimize the number of wash and incubation steps, saving you valuable time. The kit also includes unmethylated, 5-methylcytosine and 5-hydroxymethylcytosine DNA controls and PCR primers that can be used to verify the efficiency of the enrichment. Additionally, the kit contains a bar magnet for easy separation and elution of the enriched hydroxymethylated DNA. Click the Data tab below to see results from the kit; manuals can be downloaded under the Documents tab.
Documents (3)
5-hmC Info
5-hydroxymethylcytosine (5-hmC)
Methylated DNA Immunoprecipitation (MeDIP) is an immunocapture technique in which an antibody specific for methylated cytosines is used to immunoprecipitate methylated genomic DNA fragments. For hMeDIP, the antibody used for immunocapture is a 5-hydroxymethylcytidine antibody that recognizes DNA fragments containing 5-hydroxymethylcytosine (5-hmc). This selectivity is important as most common approaches to analyze DNA methylation, such as enzymatic approaches and bisulfite conversion, are unable to distinguish between 5-hmC and 5-mC.
The 5-hydroxymethylcytosine modification results from the enzymatic conversion of 5-methylcytosine into 5-hydroxymethylcytosine by the TET family of cytosine oxygenases. While the precise function of 5-hmC has yet to be determined, it has been postulated that it could represent a pathway to demethylated DNA, as 5-hydroxymethylcytosine is repaired as mismatched DNA and replaced with unmethylated cytosine.
The affinity of the antibody used in the hMeDIP kit enables detection of hydroxymethylated cytosines regardless of their context. This means that the methylation does not need to occur in the context of a CpG dinucleotide, unlike methyl-binding protein (MBD) affinity enrichment methods which require CpG methylation for proper detection. Another advantage of the hMeDIP Kit is that the 5-hmC antibody works efficiently to immunoprecipitate either double-stranded or single-stranded DNA.
hMeDIP Kit Advantages
- Specific antibody only enriches 5-hmC methylated DNA
- Works with either double-stranded or single-stranded DNA
- Optimized magnetic protocol to minimize hands-on time
- Kit includes unmethylated, 5-methylcytosine and 5-hydroxymethylcytosine control DNAs and PCR primers to verify efficiency
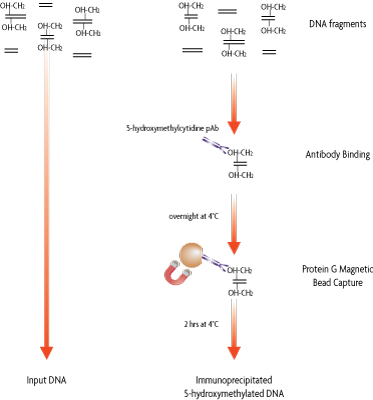
Flow Chart of hMeDIP Method
Figure 1: Flow chart of the hMeDIP Method.
Fragmented genomic DNA containing 5-hydroxymethylcytosine can be specifically captured from the rest of the genomic DNA with the purified 5-hmC antibody. Following an overnight incubation, the antibody/DNA complex is captured with protein G magnetic beads. Enriched DNA is then eluted from the beads. (Click image to enlarge.)
Data
hMeDIP
The hMeDIP Kit is able to selectively enrich for DNA fragments containing 5-hydroxymethylcytosine DNA methylation from the rest of the genomic DNA population. The hMeDIP Kit was tested using the included methylated DNA controls. The control DNAs contain a 338 base pair DNA fragment consisting of 122 cytosine residues that are completely unmethylated, completely 5-methylcytosine methylated or completely 5-hydroxymethylcytosine metylated. These DNA controls were spiked into human, male genomic DNA at a ratio of 1:20,000 (spike:genomic DNA) and run through the hMeDIP Kit. Eluted DNA was purified with Active Motif's Chromatin IP DNA Purification Kit (Catalog No. 58002) and tested in real time PCR with the included PCR primer set. Results are shown in Figure 1 that indicate the specificity of the 5-hmC antibody for detecting only the 5-hydroxymethylated DNA. Fold enrichment was calculated by normalizing the DNA enriched with the 5-hmC antibody against the DNA enriched with the negative control rabbit IgG antibody. These results are shown in Figure 2.
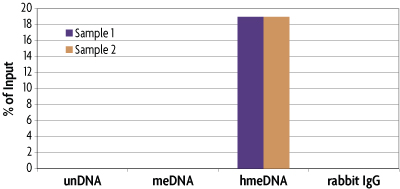
Figure 1: Specificity of the hMeDIP Kit. MseI digested human genomic DNA (500 ng) was spiked with 25 pg of either methylated, hydroxymethylated or unmethylated APC DNA. These samples were then processed using the hMeDIP Kit with the purified 5-hydroxymethylcytidine pAb. Eluted DNA was purified and tested using real time PCR with the included APC PCR primer mix. The 5-hydroxymethylcytidine antibody specifically enriched the IP sample containing the hydroxymethylated APC DNA, but did not enrich for methylated or unmethylated DNA. The APC locus analyzed in this experiment is not methylated in human genomic DNA and therefore should not amplify. This experiment was performed to detect the presence of the spiked control DNA only. Data shown are the results from samples assayed in duplicate. These results are provided for demonstration only.

Figure 2: Fold enrichment of eluted DNA from 5-hmC IP as compared to negative control IP. MseI digested human genomic DNA (500 ng) was spiked with 25 pg of either methylated, hydroxymethylated or unmethylated APC DNA. These samples were then processed using the hMeDIP Kit with the purified 5-hydroxymethylcytidine pAb. Eluted DNA was purified and tested using real time PCR with the included APC PCR primer mix. The 5-hydroxymethylcytidine antibody specifically enriched the IP sample containing the hydroxymethylated APC DNA, but did not enrich for methylated or unmethylated DNA. The APC locus analyzed in this experiment is not methylated in human genomic DNA and therefore should not amplify. This experiment was performed to detect the presence of the spiked control DNA only. The fold enrichment represents the amount of IP DNA recovered from each spike reaction with the 5-hmC antibody normalized against the negative control rabbit IgG IP reaction. Data shown are the results from samples assayed in duplicate. These results are provided for demonstration only.
Contents
Contents & Storage
The hMeDIP Kit contains a purified 5-hydroxymethylcytidine polyclonal antibody, a negative control rabbit IgG antibody, PIC, Buffer C, Buffer D, Elution Buffer AM2, Neutralization Buffer, PCR tubes, Protein G magnetic beads and a bar magnet for the separation and elution of DNA fragments containing 5-hmC. Protocols are provided for the fragmentation of sample DNA all the way through to real time PCR analysis of a locus of interest.
The kit also includes DNA controls and PCR primers. A 338 bp sequence of the APC locus that is unmethylated, fully hydroxymethylated or fully methylated is included for optional spike experiments to determine the efficiency of the immunoprecipitation. APC PCR primers are included to verify the enrichment of the DNA controls.
Storage condiitons range from room temperature to -20°C; please refer to the manual for proper storage temperatures of each component.