10 tips on how to best optimize siRNA transfection
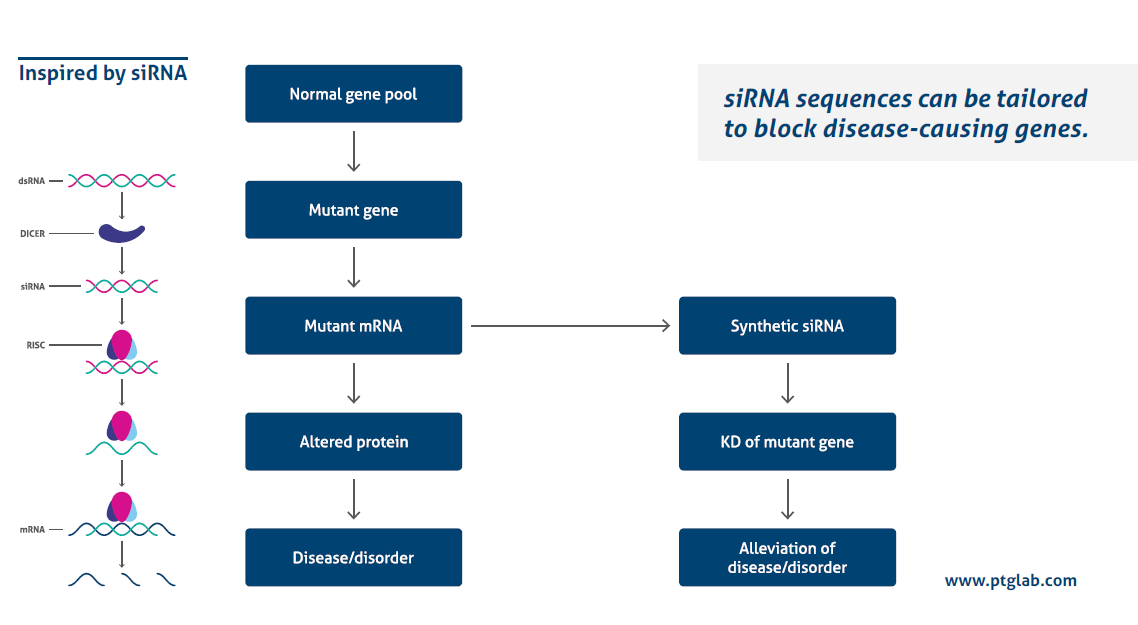
siRNA transfection is a powerful tool used to understand the mechanisms of gene regulation and molecular pathways.
How does it work?
Transfecting siRNA with a high efficiency while avoiding side effects is influenced by several different factors. The following 10 tips will help you to optimize your siRNA transfection.
1. Protocol optimization
The following factors influence your siRNA transfection and have to be optimized for every new experiment:
- Cell type
- Cell growth rate
- Cell density
- Cell viability
- Transfection method
- Quality/quantity of siRNA
- Time of transfection
2. RNase-free environment
Before starting your experiment, clean your working space with RNase-decontaminating solutions. When working with siRNA, always use gloves, changing them after touching any surface. Use pipettes with RNase-free tips and do not use these pipettes for other experiments.
3. Designing siRNA
- Around 21–23 nt in length
- G/C content: 30–50%
- No base pair mismatch
- siRNA should not bind to introns
- No sequence that shows homology to other coding sequences
4. Working with a new target/siRNA/cell line
Be prepared to run multiple test transfections in order to optimize the best conditions.
5. siRNA controls
It is essential to use suitable controls in order to correctly interpret the results.
- Positive control: A known siRNA to give a high knockdown of the target of interest.
- Negative control: A non-silencing siRNA helps to identify non-specific changes in gene expression.
- Another siRNA against the same target: A second siRNA against the same target but against another region of the mRNA will prove the specificity of the silencing effects.
- Untreated sample: The normal gene expression level should be determined using a non-transfected control.
- Transfection control siRNA: Fluorescently labeled siRNA simplifies targeting of the knockdown effect.
- Mock-transfected control: Cells transfected with the transfection reagent only, without siRNA.
6. Validation of siRNA data
- Titrate the siRNA: Use different concentrations of siRNA and consult the manufacturer’s instructions, generally a concentration of 5–100 nM is used. Use the lowest working concentration.
- Monitor RNA and protein level: Successful mRNA silencing without a corresponding decrease in protein level suggests a slow protein turnover.
7. Cell culture
Cells should be in optimal physiological condition at the time of transfection. Cells need to be passaged frequently and the transfection should always be carried out under the same culture conditions. Usually a high cell density of around 70% is needed at the time of transfection. However, this depends on the cell type and should be determined for each experiment.
8. Serum or serum-free medium?
Most transfection reagents require a serum-free medium for the initial dilution of the siRNA complex. If sera are added to the cell culture while the transfection is carried out, the quality/lot might also affect the experiment.
9. Antibiotic or antibiotic-free medium?
Dependent on the combination of cell type and transfection reagent used, cell permeability is very sensitive during a transfection. The use of antibiotics can therefore cause cell death.
10. Duration of siRNA silencing
In general, the earliest time after which the silencing effect can be observed is 24 hours. It retains cell type dependent for 4–7 days.
Futher References
D.S . Anson et al., The use of retroviral vectors for gene therapy-what are the risks? A review of retroviral pathogenesis and its relevance to retroviral vector-mediated gene delivery. Genet. Vaccines Ther. 2, 9. (2004)
F. Barthel et al, Gene transfer optimization with lipospermine-coated DNA, DNA Cell Biol. 12, 553–60( 1993).
P.L. Felgner et al, Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J. Biol. Chem. 269, 2550–61 (1994).
K. Huppi, S. Martin, et al., Defining and Assaying RNAi in Mammalian Cells. Mol. Cell. 17(1), 1-10 (2005)
D.H. Kim et al., Nature Reviews Genetics 8, 173–184 (2007)
L.P. Lim, N.C. Lau, et al., Microarray analysis shows that some microRNAs down regulate large numbers of target mRNAs. Nature. 433(7027), 769-773 (2005).
S. Loyter et al., Mechanisms of DNA uptake by mammalian cells: Fate of exogenously added DNA monitored by the use of fluorescent dyes. Proc. Natl. Acad. Sci. USA 79, 422–6. (1982)
Whither RNAi? Nat. Cell. Biol. 5(6), 489-90 (2003)




