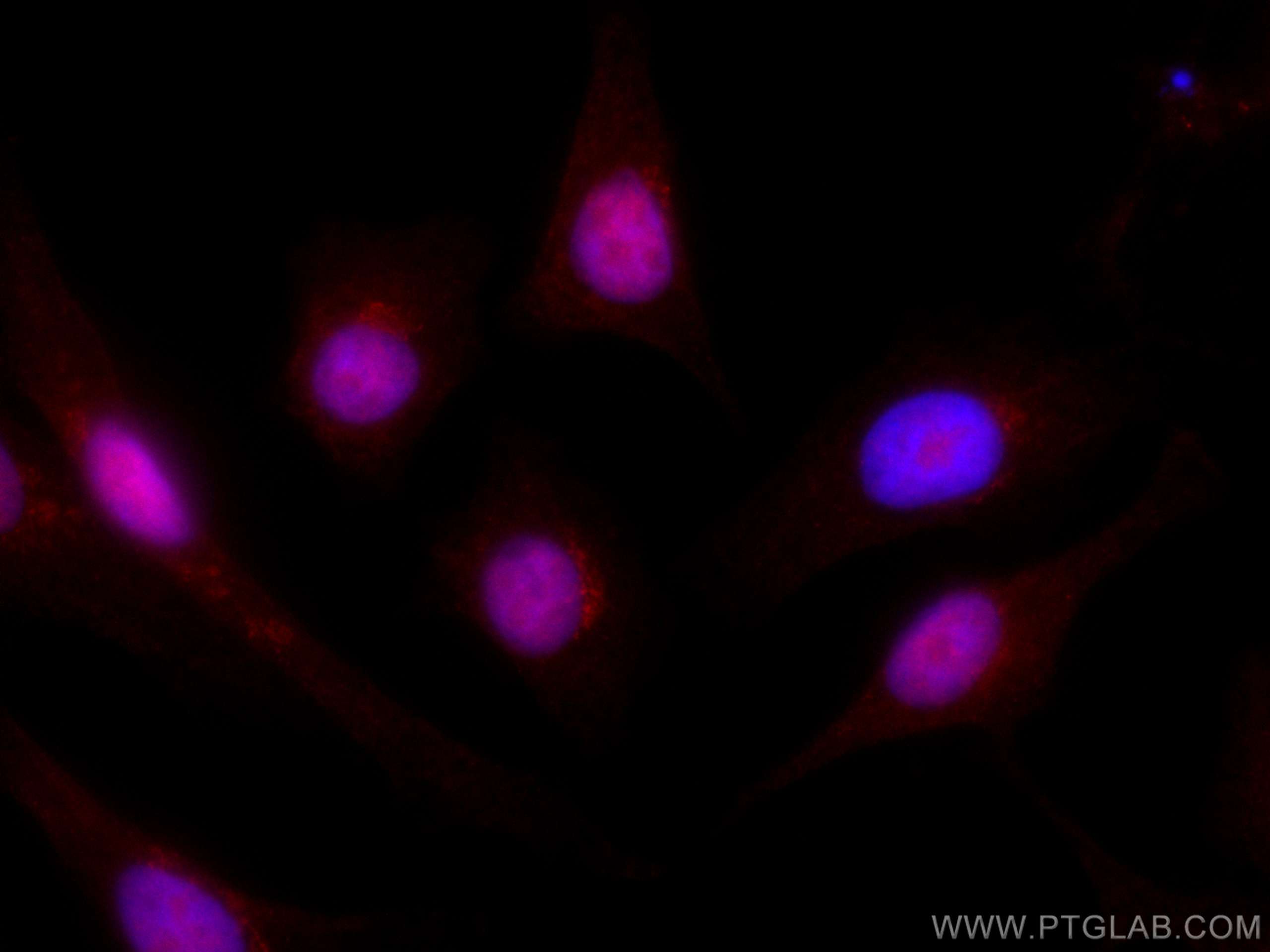
验证数据展示
经过测试的应用
| Positive IF/ICC detected in | A549 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
CL594-66941 targets RDM1 in IF/ICC applications and shows reactivity with Human samples.
| 经测试应用 | IF/ICC Application Description |
| 经测试反应性 | Human |
| 免疫原 | RDM1 fusion protein Ag28686 种属同源性预测 |
| 宿主/亚型 | Mouse / IgG2a |
| 抗体类别 | Monoclonal |
| 产品类型 | Antibody |
| 全称 | RAD52 motif 1 |
| 别名 | RAD52 homolog B, RAD52 motif 1, RAD52B, RDM1 |
| 计算分子量 | 284 aa, 32 kDa |
| 观测分子量 | 32 kDa |
| GenBank蛋白编号 | BC038301 |
| 基因名称 | RDM1 |
| Gene ID (NCBI) | 201299 |
| RRID | AB_2920061 |
| 偶联类型 | CoraLite®594 Fluorescent Dye |
| 最大激发/发射波长 | 588 nm / 604 nm |
| 形式 | Liquid |
| 纯化方式 | Protein A purification |
| UNIPROT ID | Q8NG50 |
| 储存缓冲液 | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA , pH 7.3 |
| 储存条件 | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
RDM1, also named as RAD52B, encodes a RNA recognition motif (RRM)-containing protein confered resistance to the antitumor agent cisplatin. The expression of RDM1 is closely related to the cell proliferation and apoptosis of thyroid carcinoma cells and RDM1 may be a potential marker of lung adenocarcinoma. RDM1 has many isoforms (8 kDa, 12-19 kDa and 25-32 kDa) with long or short N-terminus and a great diversity in the exonic composition of C-terminus generated by alternative pre-mRNA splicing and differential usage of two translational start sites. RDM1 protein can be expressed differently in cytoplasm or nuclear/nucleolar in the absence of stress, proteotoxic stress and mild heat shock, suggesting functional differences among the RDM1 proteins. ( PMID: 28939762, 17905820, 28939762, 30069034)
实验方案
| Product Specific Protocols | |
|---|---|
| IF protocol for CL594 RDM1 antibody CL594-66941 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |