验证数据展示
经过测试的应用
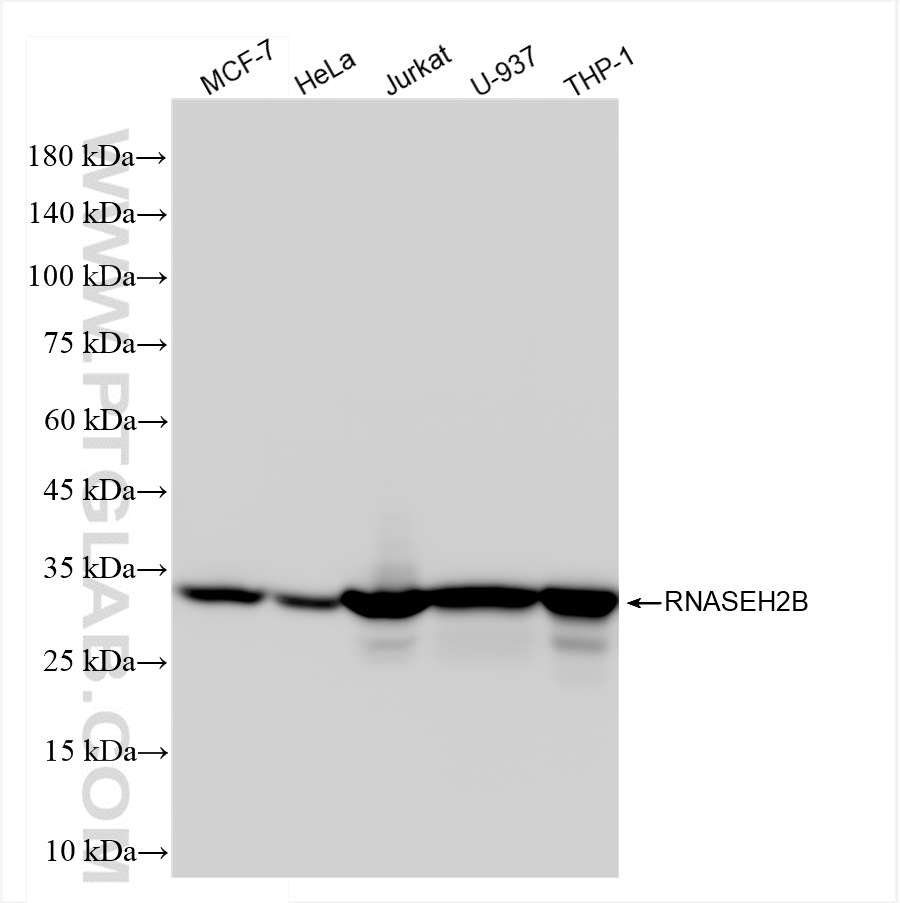
| Positive WB detected in | MCF-7 cells, HeLa cells, Jurkat cells, U-937 cells, THP-1 cells |
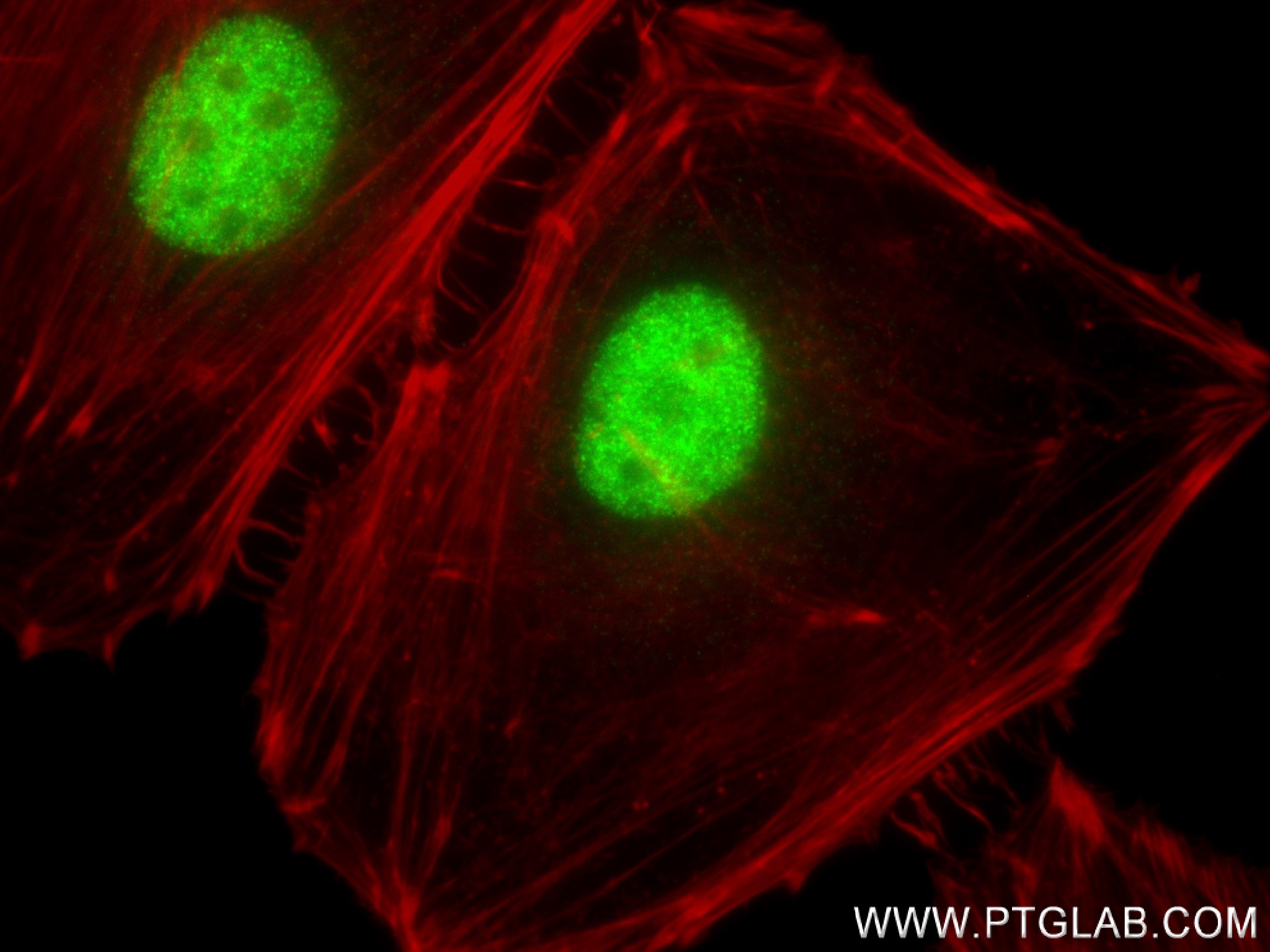
| Positive IF/ICC detected in | HeLa cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:6000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
84097-6-RR targets RNASEH2B in WB, IF/ICC, ELISA applications and shows reactivity with human samples.
| 经测试应用 | WB, IF/ICC, ELISA Application Description |
| 经测试反应性 | human |
| 免疫原 | RNASEH2B fusion protein Ag33669 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Recombinant |
| 产品类型 | Antibody |
| 全称 | ribonuclease H2, subunit B |
| 别名 | ribonuclease H2, subunit B, Ribonuclease H2 subunit B, DLEU8, Aicardi-Goutieres syndrome 2 protein, AGS2 |
| 计算分子量 | 331 aa, 37 kDa |
| 观测分子量 | 35 kDa |
| GenBank蛋白编号 | BC036744 |
| 基因名称 | RNASEH2B |
| Gene ID (NCBI) | 79621 |
| RRID | AB_3671663 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Protein A purfication |
| UNIPROT ID | Q5TBB1 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
Ribonuclease H2 is the major nuclear enzyme degrading cellular RNA/DNA hybrids in eukaryotes and the sole nuclease known to be able to hydrolyze ribonucleotides misincorporated during genomic replication. RNASEH2B and RNASEH2C are the non-catalytic subunits of RNase H2 complex. Defects in RNASEH2B are a cause of Aicardi-Goutieres syndrome type 2 (AGS2), a genetically heterogeneous disease with clinical features as thrombocytopenia, hepatosplenomegaly and elevated hepatic transaminases along with intermittent fever may erroneously suggest an infective process.
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for RNASEH2B antibody 84097-6-RR | Download protocol |
| IF protocol for RNASEH2B antibody 84097-6-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |