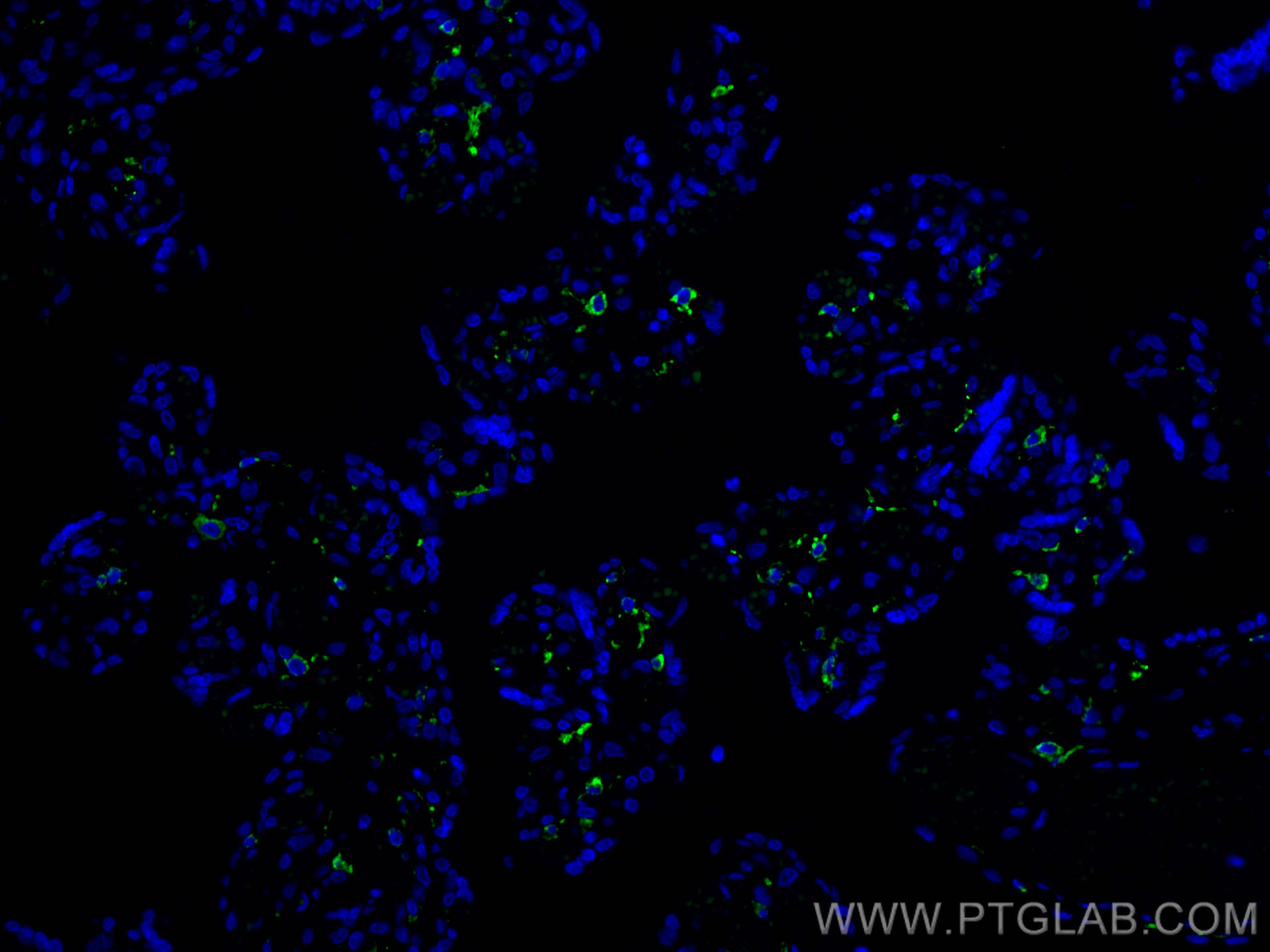
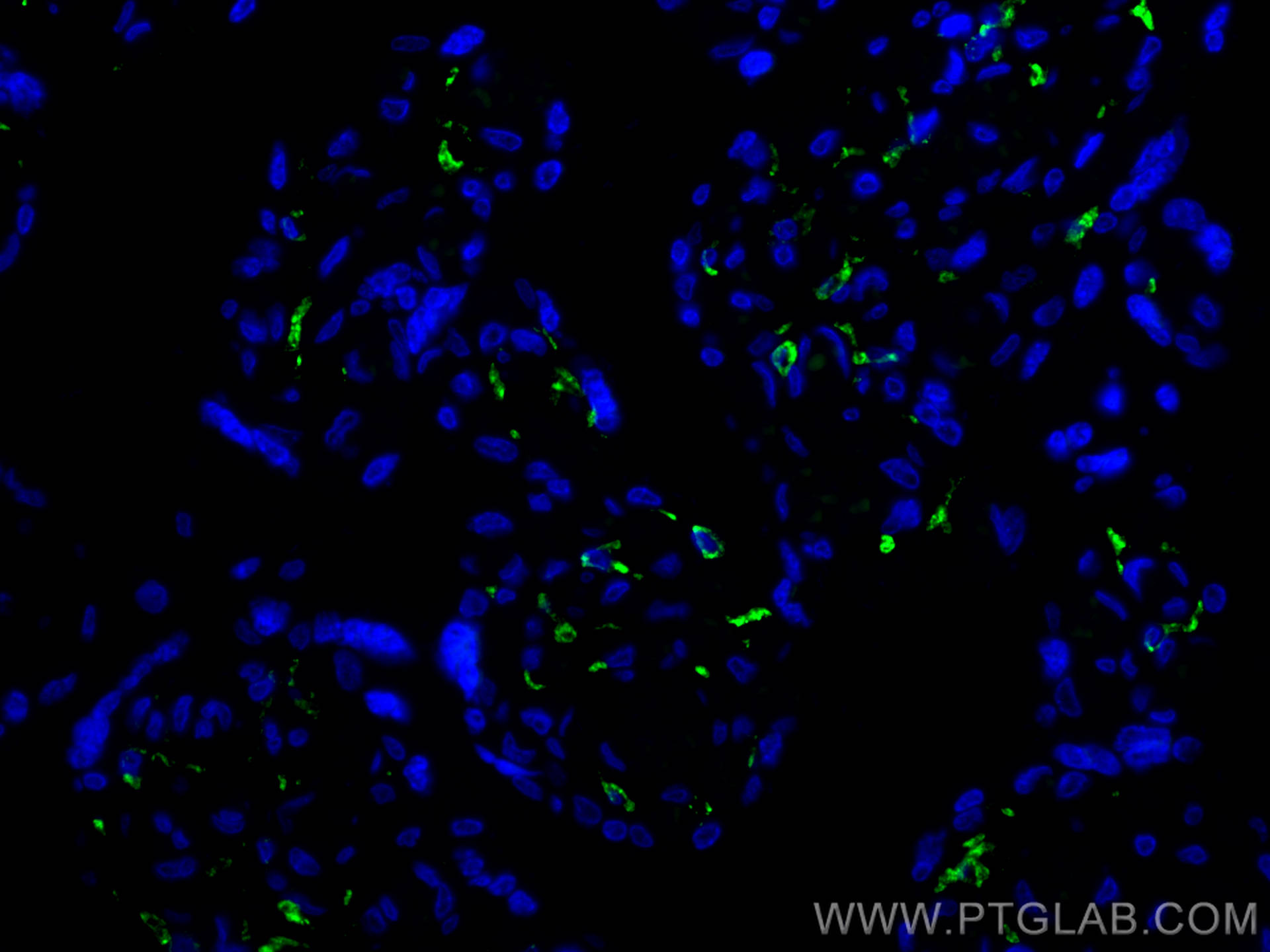
验证数据展示
经过测试的应用
| Positive IF-P detected in | human placenta tissue |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Immunofluorescence (IF)-P | IF-P : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
CL488-81525 targets CD206 in IF-P applications and shows reactivity with Human, Pig samples.
| 经测试应用 | IF-P Application Description |
| 经测试反应性 | Human, Pig |
| 免疫原 | Peptide 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Recombinant |
| 产品类型 | Antibody |
| 全称 | mannose receptor, C type 1 |
| 别名 | CD206, CLEC13D, Macrophage mannose receptor 1, mannose receptor, C type 1, MMR, MRC1 |
| 计算分子量 | 166 kDa |
| 观测分子量 | 170 kDa |
| GenBank蛋白编号 | NM_002438 |
| 基因名称 | CD206 |
| Gene ID (NCBI) | 4360 |
| RRID | AB_3673047 |
| 偶联类型 | CoraLite® Plus 488 Fluorescent Dye |
| 最大激发/发射波长 | 493 nm / 522 nm |
| 形式 | Liquid |
| 纯化方式 | Protein A purification |
| UNIPROT ID | P22897 |
| 储存缓冲液 | PBS with 50% Glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| 储存条件 | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
CD206, also named as MMR, CLEC13D and MRC1, is a type I membrane receptor that mediates the endocytosis of glycoproteins by macrophages. CD206 has been shown to bind high-mannose structures on the surface of potentially pathogenic viruses, bacteria, and fungi so that they can be neutralized by phagocytic engulfment. CD206 is a 170 kDa transmembrane protein which contains 5 domains: an amino-terminal cysteine-rich region, a fibronectin type II repeat, a series of eight tandem lectin-like carbohydrate recognition domains (responsible for the recognition of mannose and fucose), a transmembrane domain, and an intracellular carboxy-terminal tail. It is expressed on most tissue macrophages, in vitro derived dendritic cells, lymphatic and sinusoidal endothelia.
实验方案
| Product Specific Protocols | |
|---|---|
| IF protocol for CL Plus 488 CD206 antibody CL488-81525 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |