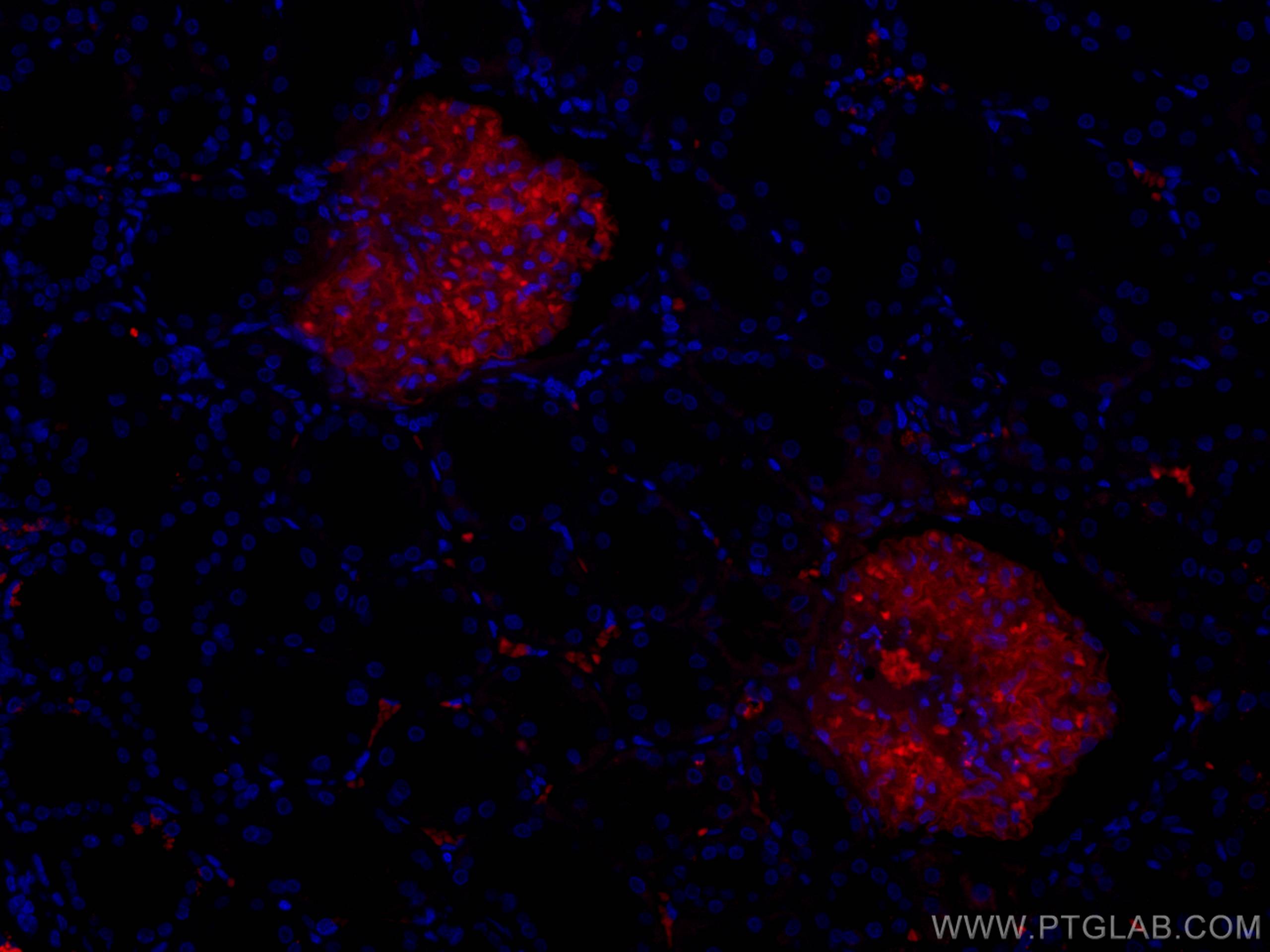
验证数据展示
经过测试的应用
| Positive IF-P detected in | human kidney tissue |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Immunofluorescence (IF)-P | IF-P : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
CL594-67000 targets PTPRO in IF-P applications and shows reactivity with Human, Pig samples.
| 经测试应用 | IF-P Application Description |
| 经测试反应性 | Human, Pig |
| 免疫原 | PTPRO fusion protein Ag8284 种属同源性预测 |
| 宿主/亚型 | Mouse / IgG2b |
| 抗体类别 | Monoclonal |
| 产品类型 | Antibody |
| 全称 | protein tyrosine phosphatase, receptor type, O |
| 别名 | GLEPP1, PTP U2, PTPase U2, PTPRO, PTPU2, R PTP O |
| 计算分子量 | 138 kDa |
| 观测分子量 | 160 kDa, 180-220 kDa |
| GenBank蛋白编号 | BC035960 |
| 基因名称 | PTPRO |
| Gene ID (NCBI) | 5800 |
| RRID | AB_2920069 |
| 偶联类型 | CoraLite®594 Fluorescent Dye |
| 最大激发/发射波长 | 588 nm / 604 nm |
| 形式 | Liquid |
| 纯化方式 | Protein A purification |
| UNIPROT ID | Q16827 |
| 储存缓冲液 | PBS with 50% Glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| 储存条件 | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
PTPRO(receptor-type tyrosine-protein phosphatase O) is also named as GLEPP1, PTPU2 and belongs to the protein-tyrosine phosphatase family. The protein is a receptor-like membrane protein tyrosine phosphatase expressed at the apical membrane of the podocyte foot processes in the kidney(PMID:21722858). The 1,159-amino acid predicted mature protein contains a large extracellular domain, a single transmembrane domain, and a single intracellular PTPase domain. Defects in PTPRO are the cause of nephrotic syndrome type 6 (NPHS6). In reducing conditions, PTPRO can form a smear from 180 to 220 kDa; In non-reducing conditions, a band appeares around 350 kDa, which likely represents the dimeric form of full-length PTPRO (PMID: 19573017).
实验方案
| Product Specific Protocols | |
|---|---|
| IF protocol for CL594 PTPRO antibody CL594-67000 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |