mRNA Processing
RNA processing is important for the post-transcriptional regulation of gene expression.
Introduction
RNA processing is important for the post-transcriptional regulation of gene expression, and is a rate-limiting step in the expression of proteins. The majority of RNA processing occurs simultaneously with transcription, at which stage several processes shape the nascent messenger RNA (mRNA) molecule: mRNA capping, splicing, editing and 3’ end formation. Throughout these processes, various enzymes and proteins play an important role in stabilizing mRNA transcripts.
In addition to co-transcriptional regulation, RNAs – both coding mRNAs and non-coding species – can also be controlled at the posttranscriptional level. There are around 150 known RNA modifications, yet our knowledge about their occurrence and function is still limited. One thing is certain though: post-transcriptional regulation of gene expression is much more intricate than previously thought. Unravelling the inner workings of it is essential to understanding gene expression in its entirety, and will provide valuable insights into post-transcriptional dysfunction in numerous genetic diseases and cancer. This catalog contains antibodies targeting proteins involved in several aspects of RNA processing, selected for researchers making the discoveries in this area.
CIRBP
Cellular stress, such as a sudden drop in temperature, results in the expression of several proteins involved in transcription, translation and splicing of cold-shock proteins. Cold-inducible mRNA binding protein, CIRBP, (also named A18HNRNP or CIRP) plays an important role in the protection of gene transcripts from genotoxic stressors, such as hypoxia, hypothermia and UV light. When cells are subjected to stress, CIRBP has the ability to translocate from the nucleus to the cytoplasm where it acts to stabilize mRNA.
Recently, CIRBP has been linked with oral squamous cell carcinoma (OSCC); the commonest type of head and neck malignancy, representing almost 95% of head and neck cancers. In a recent study, Ren et al. showed elevated expression of CIRBP and toll-like receptor 4 (TLR4) in OSCC patients.
Previously, CIRBP has been shown to induce inflammatory action through TLR4.The development of OSCC is due, in part, to chronic inflammation, although a full understanding of the pathogenesis of the condition still remains elusive. It has been suggested that activation of TLR4 can induce chronic inflammation, which, in turn, promotes tumor growth.
Using Proteintech’s antibody against CIRBP (10209-2-AP), Ren and colleagues selected biopsy samples from OSCC patients and control subjects and analyzed them for CIRBP and TLR4 expression. They were able to demonstrate that CIRBP was overexpressed in 37.7 percent of patients and under-expressed in a further 62.3 percent. On the contrary, tissue from control patients showed no expression of TLR4 or CIRBP. Further to this, the authors sought to compare the relationship between survival rates of patients with OSCC and overexpression of CIRP or TLR4. It was shown that overexpression of either marker resulted in a significantly poorer prognosis. This could be partly due to CIRBP’s role in the underlying pathogenesis of the OSCC cases from the outset, but it has also been reported that CIRBP protects against TNF-alpha induced apoptosis. Thus, it could also confer OSCC’s with a survival advantage.
The findings from this study support the hypothesis that CIRBP and/or TLR4 may play in important biological function in the development of OSCC. Furthermore, they may in future serve as diagnostic markers in predicting patient outcomes.
| Antibody | Catalog number | Type | Applications |
| CIRBP | 10209-2-AP | Rabbit Polyclonal | ELISA, IF, IHC, IP, Supershift Assay, WB |
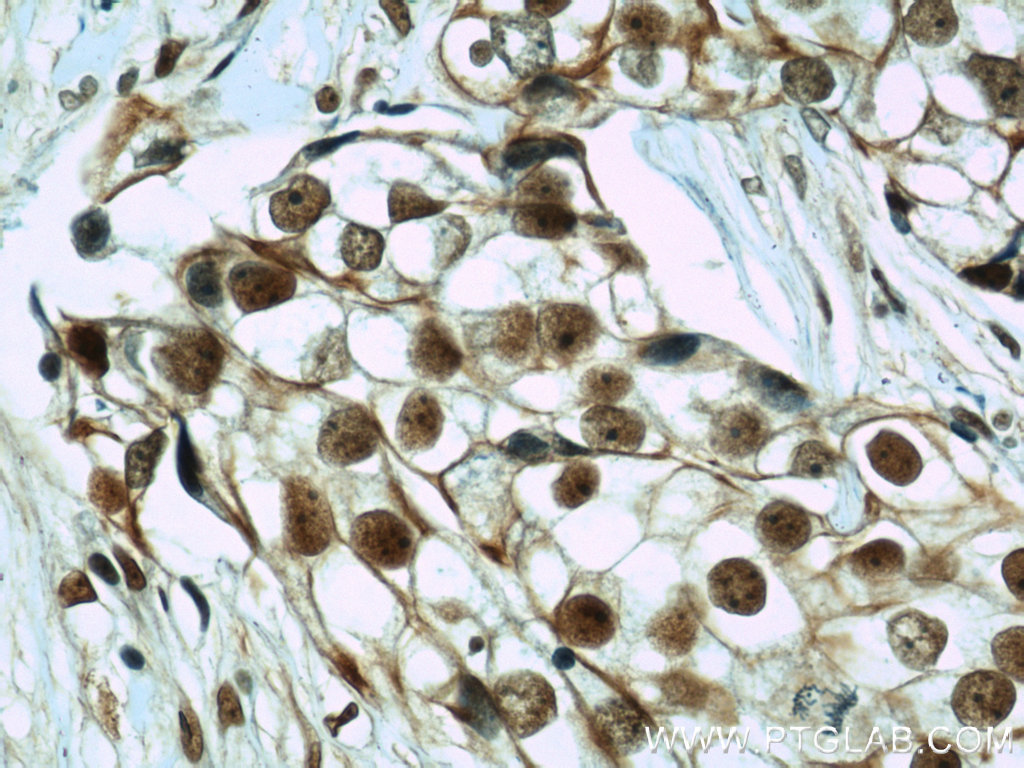
Right: Immunohistochemical of paraffin-embedded human breast cancer using CIRBP antibody (10209-2-AP) at a dilution of 1:50 (40x objective).
RTCB
The endoplasmic reticulum (ER) is an organelle found in eukaryotic cells in two forms: smooth ER and rough ER. The two types of ER function primarily to aid the folding of proteins and the transport of those newly synthesized to the Golgi apparatus. The maintenance of protein folding networks found within the ER is carried out by the unfolded protein response (UPR). The UPR consists of three signaling branches distinguished by the principal enzymes by which they act: AFT6, PERK and IRE1. The IRE1 branch of the UPR initiates the splicing of mRNA of XBP1 during ER stress. XBP1 is a key UPR transcription factor.
During splicing, the enzyme known as IRE1- alpha cleaves the sole intron of the XBP1 transcript, leaving two remaining exons needing ligation. However, the identity of the enzyme that does this was a mystery until the publication of a Molecular Cell paper in September 2014.
Using Proteintech’s anti-RtcB antibody (19809-1-AP), the Lu et al. Molecular Cell paper showed that RtcB is the elusive RNA ligase essential for the joining of XBP1 exons during the UPR. The authors were able to show that RctB acts downstream of IRE1- alpha, demonstrating this occurs after removal of intron sequences. Furthermore, XBP1 mRNA splicing is defective in RtcB knockout cells during times of ER stress, which is restored following genetic rescue.
The authors have successfully demonstrated that RtcB is the ligase responsible for splicing occurring during UPR, a pathway now considered to have great importance in neurodegenerative disease.
| Antibody | Catalog number | Type | Applications |
| RTCB | 19809-1-AP | Rabbit Poly | ELISA, WB, IP, IF |
SF3B3
During mRNA synthesis, mRNA splicing removes introns from nuclear pre-mRNA in a two-step process. This process occurs in a ribonucleoprotein complex known as the spliceosome, and requires the interaction of several proteins such as Splicing factor 3b (SF3B3). Along with splicing factor 3A and a 12s RNA unit, SF3B3 makes up the U2 small nuclear ribonucleoproteins complex (U2 snRNP), which is recruited along with other U snRNPs to a pre-mRNA substrate to assemble spliceosomes. These undergo ATP-dependent rearrangements to form intermediates known as E, A, B, Bact, B*, and C complexes. Not all of these interactions have been characterized, partly because it has not been possible to arrest the complex at clearly defined chemical states.
In a recent publication, Ilagan et al. devised a way of stalling human spliceosomes in a new post-catalytic state (which they named the P complex). Quantitative comparisons of P complex and C complex units showed an increase of SF3b components in the spliceosome and a loss of putative RNAdependent ATPase DHX35. Using Proteintech’s anti-SF3B3 antibody (14577-1-AP) and their novel method of spliceosome arrest, the authors have taken a snapshot of the human spliceosome before and after splicing. The work of Ilgan et al. has opened a new window to viewing rearrangements near the active site of spliceosomes, allowing us to peer into the inner workings of exon ligation and mRNA release.
| Antibody | Catalog number | Type | Applications |
| SF3B3 | 14577-1-AP | Rabbit Poly | ELISA, WB, IP, IHC, IF |
More related antibodies
| Antibody | Catalog number | Type | Applications |
| DHX35 | 24923-1-AP | Rabbit Poly | ELISA, WB, IHC |
| SF3A1 | 15858-1-AP | Rabbit Poly | ELISA, WB, IP, IHC, IF |
| SF3A2 | 15596-1-AP | Rabbit Poly | ELISA, WB, IHC |
| SF3A3 | 12070-1-AP | Rabbit Poly | ELISA, IF, IHC, IP, WB |
METTL3
Methyltransferase like 3, also known as METTL3, is an enzyme involved in the posttranscriptional methylation of adenosine bases in mRNA, with its action resulting in the formation of N6 methyladenosine (m6A). This form of modification is the most prevalent internal modification in mammalian mRNA and non-coding RNA: RNA-immunoprecipitation methods followed by next generation sequencing have uncovered that more than 7,000 mRNAs and over 2,000 long non- coding RNAs contain m6A sites and these are most often located at stop codons.³ The precise biological functions of m6A-methyltransferases are not fully characterized, but evidence so far implicates a role in embryo development, gametogenesis and stem cell self-renewal.
METTL3 also interacts with METTL14, another enzyme involved in the formation of m6A. The interaction between METTL3 and METTL14 occurs in the form of a heterodimer complex that results in the m6A depositions on nuclear RNA. The paper that reported this finding also featured a Proteintech antibody recognizing METTL3 (15073-1-AP). Using gel filtration, Liu et al. were able to show that METTL3 and METTL14 formed a stable heterodimeric complex in vitro. They further demonstrated that RNA interference knockdown of METTL3, METTL14 and WTAP (Wilms Tumor 1-Associating Protein) in HeLa cells reduced m6A additions to RNA by 30%, 40%, 50% respectively.⁴ Additional investigation showed that both METTL3 and METTL14 exhibited methyltransferase activity with METTL14 showing considerably higher enzymatic activity than METTL3.
In summary, the authors showed that the newly discovered methyltransferase METTL14 forms a stable complex with METTL3, which acts to deposit m6A on RNA, with WTAP interacting with the METTL3- METTL14 complex modulating cellular m6A deposition. Furthermore, this activity is modulated by WTAP. The elucidation of this novel methyltransferase complex may prove a novel target in modulating gene expression and help shed light on the purpose of mRNA methylation in health and disease.
| Antibody | Catalog number | Type | Applications |
| METTL3 | 15073-1-AP | Rabbit poly | ELISA, IF, IP, WB (siRNA), WB |


