The highly adaptive cytoskeleton
The cytoskeleton is a highly important structure present in the cytoplasm of all cells. Learn how this is integral for many cellular functions, and the many ways researchers can study the cytoskeleton in our blog.
Written by Katarzyna Szymanska-De Wijs, PhD student at Hannover Medical School
The highly adaptive cytoskeleton
On hearing the term “cytoskeleton,” you might imagine a rigid, bone-like structure that gives shape and support to the cells. This is both right and wrong at the same time! While indeed providing spatial organization to a eucaryotic cell, the cytoskeleton constantly changes in response to cell needs and external factors. The cytoskeletal structures assemble and disassemble all the time, making the cytoskeleton one of the busiest and most dynamic parts of a cell. The basic building blocks of the cytoskeleton are the proteins actin and tubulin. Actin monomers polymerize, either in a long-range or branched manner, forming filaments, while tubulin assembles into microtubules used for the transport of intracellular cargo. Intermediate filaments, like vimentin, are the third component of the cytoskeletal network. The design of the polymer structures is highly regulated by different classes of proteins, e.g., polymerases or crosslinkers, which respond instantly to intracellular needs, as well as external danger cues [1].
Cytoskeletal rearrangements in the life of a cell
Being in constant motion translates to various activities in a cell. One of these is the involvement in the cell cycle. During mitosis, microtubules rearrange themselves into a mitotic spindle, a crucial structure that first attaches to duplicated chromosomes and then divides them into two new cells [2]. However, even before that happens, a cell that is preparing for division goes through a major change in shape. The cortical actin network slowly thins due to the increasing activity of the RhoA protein, a member of Rho-GTPases, which are considered master regulators of the cytoskeleton. This leads to cell rounding, a process crucial for mitotic spindle functions [3] (Fig. 1).
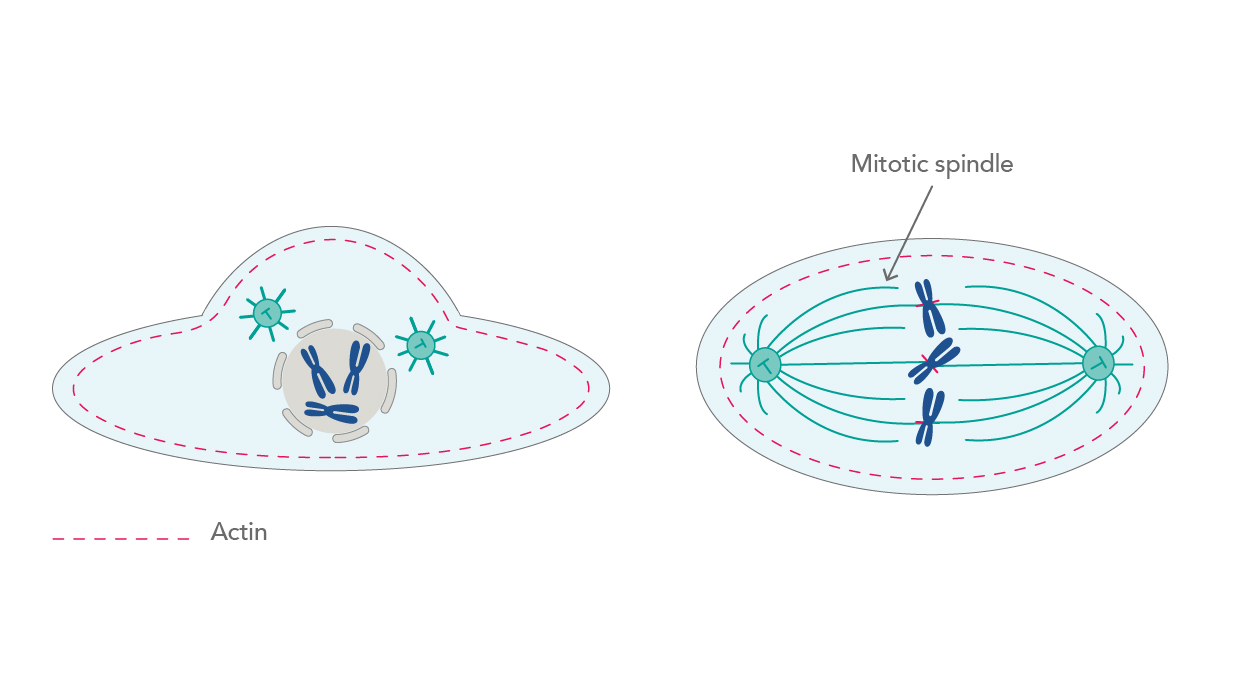 |
| Figure 1. Thinning of the actin network leads to cell rounding and promotion of mitosis. |
Motility is another cellular activity, for which the cytoskeleton is essential – for example, during leukocyte surveillance. The constant assembling and disassembling of cytoskeletal remodeling generates forces that allow cells to move and change shape. Steadily formed branched networks of filaments create membrane protrusions that promote movement at the leading edge of migrating cells as a response to chemotactic molecules sensed by the cell-surface receptors. The cell then attaches the leading site to the surface via focal adhesions, detaches the trailing edge, and uses the generated forces to move forward (Fig. 2) [4,5].
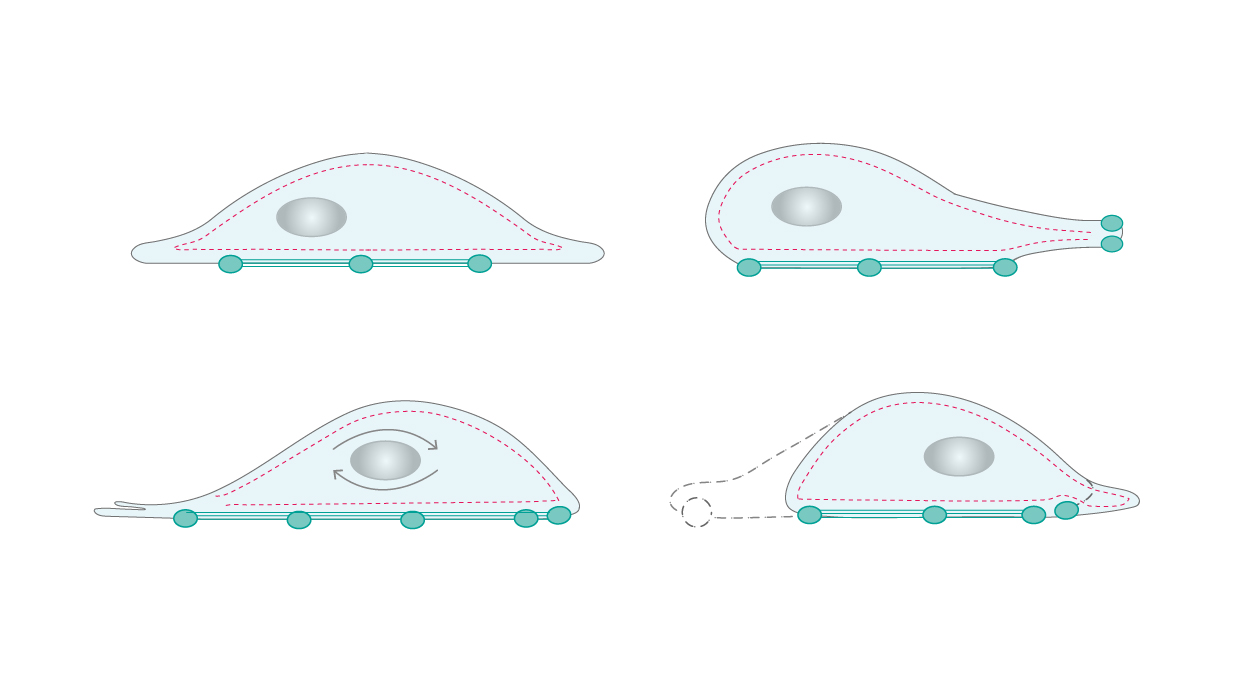 |
| Figure 2. Cellular motility requires constant assembling and disassembling of the cytoskeleton networks. |
Another important use of the cytoskeletal networks is in the transport of intracellular cargo. Motor proteins, such as myosin and kinesin, carry vesicles and other proteins to the desired destination along filaments and microtubules. This process can be controlled or disrupted by pathogens.
Intracellular Highwaymen – what happens with the cytoskeleton during infection?
Obligate intracellular pathogens establish their infection through a plethora of steps and stages – in brief, they enter a cell, replicate, and disseminate to infect the next cells. In this scenario, the first interaction with the cytoskeleton occurs right at the start, with the host cell entry of the pathogen. Bacteria, for instance, have developed three different approaches for entering mammalian cells – zipper, trigger, and phagocytosis, all of which act by activating kinase signaling cascades, actin ruffling, and internalizing the pathogen at the attachment site [6] (Fig. 3).
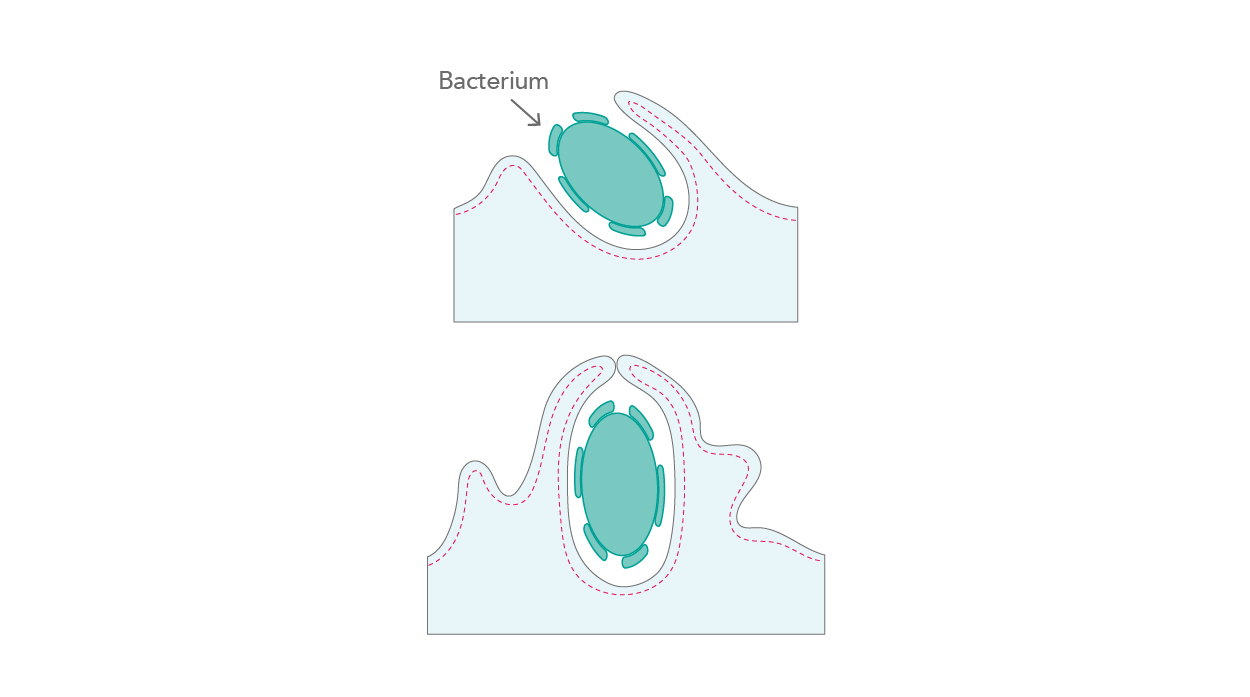 |
| Figure 3. Actin ruffling and pathogen internalization. |
Viruses have two major routes by which they enter host cells – endocytic and non-endocytic. While endocytosis allows for the easy incorporation of pathogens into the cell interior, non-endocytic entry occurs through fusion with the plasma membrane and direct penetration (Fig. 4). All these virus-driven rearrangements occur in a very short space of time; an influenza particle can enter a cell within a second, and this occurs even faster with AAV-2 particles [7].
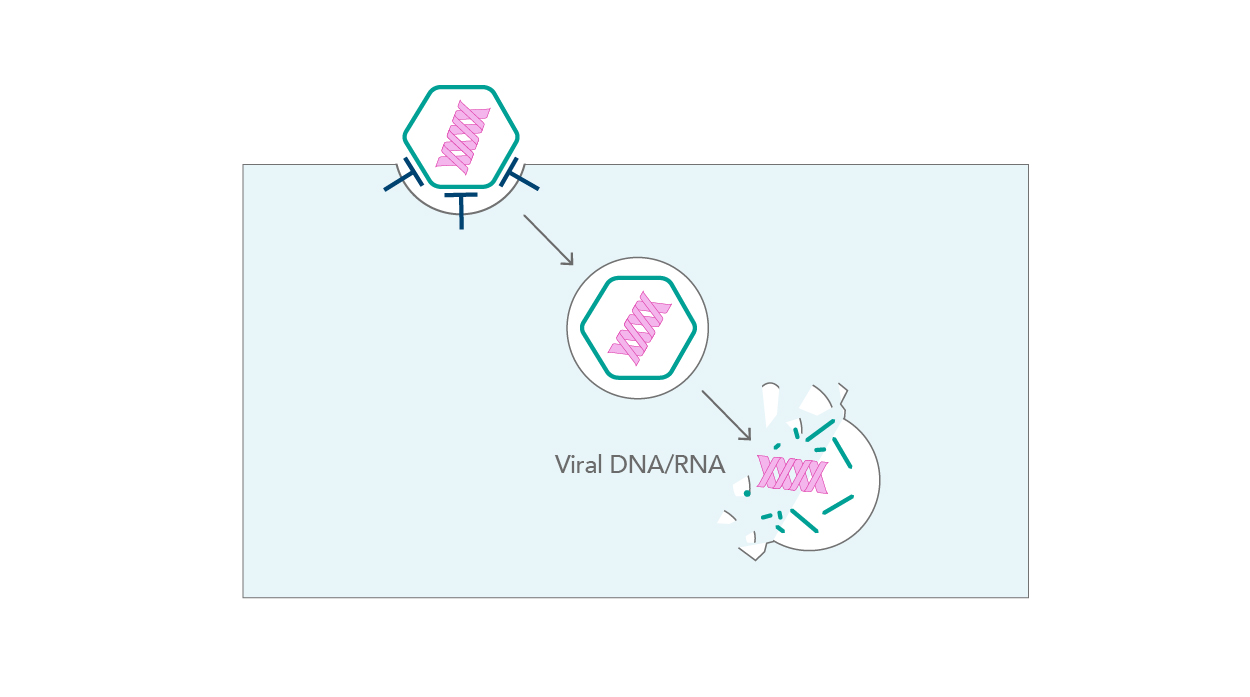 |
| Figure 4. Viral entry |
Once inside the cell, a new level of cytoskeleton manipulation occurs in order to best evade the immune response and promote infection. In some cases, bacteria build actin “tails” to promote their own mobility (e.g., Rickettsia), while others encapsulate themselves in inclusions made of actin, microtubules, and intermediate filaments (e.g., Chlamydia) [6]. Viruses, in turn, may use the cytoskeletal networks for transport through the cell by hijacking motor proteins (e.g., HSV-1) [8] or stimulating actin polymerization behind the virion (Vaccinia virus) [9,10]. Some viruses, such as the Rubella virus, completely disrupt the organization of the actin cytoskeleton to facilitate infection [10]. Recent findings have shown that even SARS-CoV-2, the culprit behind the ongoing Covid-19 pandemic, leads to dramatic cytoskeleton modifications [11]. While cortical actin accumulates at the plasma membrane to promote viral release, intermediate filaments remodel to support the formation of viral replication organelles.
When it comes to exiting the cell, pathogens enforce actin remodeling into extrusions that eventually bud off the cell. Viruses also spread through “intercellular extensions” (Fig. 5). These are actin-based structures formed by an infected cell in order to reach out to a naive cell and spread virions, as well as only partially assembled particles or sole nucleocapsids, without releasing them to the extracellular matrix [12].
 |
| Figure 5. Intercellular extensions increase the efficiency and speed of infection |
Using antibodies to study cytoskeletal rearrangements
Fluorescence imaging is undoubtedly a vital technique for deepening knowledge and understanding of the cytoskeleton. The current technology enables the detection of temporal and special patterns of cytoskeletal protein expression. The field offers different imaging modalities (e.g., confocal microscopy), as well as various approaches to sample preparation. Antibodies are the most commonly used tool for staining the desired proteins. There are two types of immunostaining – direct, with primary antibodies conjugated to a fluorophore, and indirect, where the use of secondary antibodies is necessary. The primary antibodies commonly used in Cytoskeleton research are shown in the table below, with more Cytoskeleton related antibodies here.
Table 1. Primary antibodies commonly used in Cytoskeleton research.
Proteintech provides primary antibodies labeled with CoraLite Florescent dyes. Directly conjugated antibodies offer excellent benefits, from shortening the IF workflow to reducing signal background. Discover more about the advantages of using CoraLite series products here and visit our website for more CoraLite conjugated cytoskeleton markers.
|
CoraLite®488-conjugated Beta Tubulin Monoclonal antibody
|
Table 2. Examples of CoraLite conjugated cytoskeletal antibodies.
 |
| Figure 6. Alpha-tubulin staining of HeLa cells using CL594-66031. Immunofluorescent analysis of (4% PFA) fixed HeLa cells using the CoraLite®594-conjugated alpha Tubulin antibody, at dilution of 1:100. |
Proteintech also offers recombinant antibodies to boost your Cytoskeleton research. Recombinant monoclonal antibodies are engineered using specific DNA sequences and high-yield expression systems. They display great advantages when compared to monoclonal antibodies made using hybridoma technology, such as lot-to-lot consistency, high reproducibility, and extraordinary specificity. Check out our smooth muscle actin specific recombinant antibody and learn more about recombinant antibodies here.
Phalloidin is another useful tool for staining actin filaments. As a toxin produced by the mushroom Amanita phalloides,it binds specifically to F-actin and prevents its depolymerization. Phalloidin provides much denser staining than antibodies and has very low background. Proteintech provides phalloidin conjugated to two fluorophores: CoraLite®488-Phalloidin (green) and CoraLite594-Phalloidin (red).
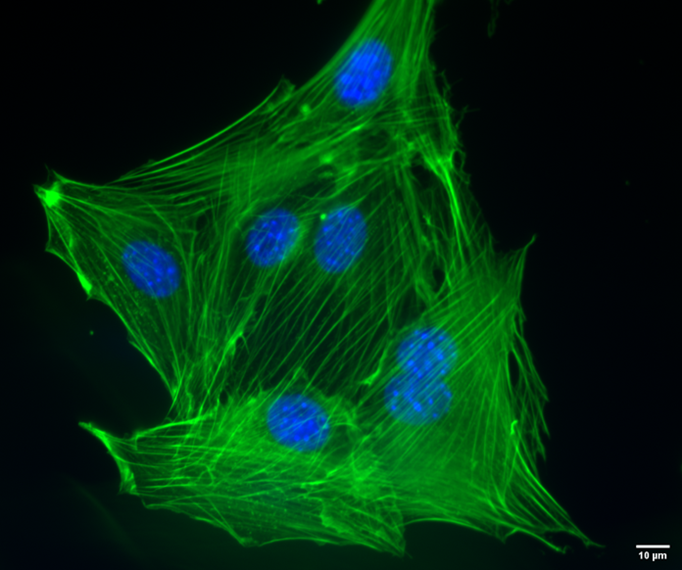 |
| Figure 7. NIH3T3 mouse fibroblasts stained with CoraLite488-Phalloidin (PF00001), used at a 1:50 dilution. Nuclei are stained with Hoechst 33342. Scale bar is 10 um. Images taken by Katarzyna Szymanska-De Wijs from Hannover Medical School. |
References:
- Fletcher, Daniel A, and R Dyche Mullins. “Cell mechanics and the cytoskeleton.” Nature vol. 463,7280 (2010): 485-92. doi:10.1038/nature08908
- Nakaseko, Y, and M Yanagida. “Cell biology. Cytoskeleton in the cell cycle.” Nature vol. 412,6844 (2001): 291-2. doi:10.1038/35085684
- Rizzelli, Francesca et al. “The crosstalk between microtubules, actin and membranes shapes cell division.” Open biology vol. 10,3 (2020): 190314. doi:10.1098/rsob.190314
- https://home.uni-leipzig.de/pwm/web/?section=introduction&page=motility accessed 22.08.2021
- Mastrogiovanni, Marta et al. “Coordinating Cytoskeleton and Molecular Traffic in T Cell Migration, Activation, and Effector Functions.” Frontiers in cell and developmental biology vol. 8 591348. 21 Oct. 2020, doi:10.3389/fcell.2020.591348
- Colonne, Punsiri M et al. “Hijacking Host Cell Highways: Manipulation of the Host Actin Cytoskeleton by Obligate Intracellular Bacterial Pathogens.” Frontiers in cellular and infection microbiology vol. 6 107. 22 Sep. 2016, doi:10.3389/fcimb.2016.00107
- Dimitrov, D. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol 2, 109–122 (2004). doi.org/10.1038/nrmicro817
- Radtke, Kerstin et al. “Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell.” Cellular microbiology vol. 8,3 (2006): 387-400. doi:10.1111/j.1462-5822.2005.00679.x
- Cudmore, S et al. “Actin-based motility of vaccinia virus.” Nature vol. 378,6557 (1995): 636-8. doi:10.1038/378636a0
- Cudmore, S et al. “Viral manipulations of the actin cytoskeleton.” Trends in microbiology vol. 5,4 (1997): 142-8. doi:10.1016/S0966-842X(97)01011-1
- Cortese, Mirko et al. “Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies.” Cell host & microbe vol. 28,6 (2020): 853-866.e5. doi:10.1016/j.chom.2020.11.003
- Cifuentes-Muñoz, Nicolás et al. “Direct cell-to-cell transmission of respiratory viruses: The fast lanes.” PLoS pathogens vol. 14,6 e1007015. 28 Jun. 2018, doi:10.1371/journal.ppat.1007015
- All figures executed by Frederik de Wijs


